Rational engineering of enzyme allosteric regulation through sequence evolution analysis
- PMID: 22807670
- PMCID: PMC3395594
- DOI: 10.1371/journal.pcbi.1002612
Rational engineering of enzyme allosteric regulation through sequence evolution analysis
Abstract
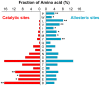
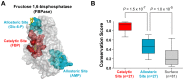
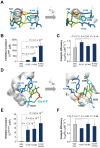
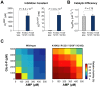
Control of enzyme allosteric regulation is required to drive metabolic flux toward desired levels. Although the three-dimensional (3D) structures of many enzyme-ligand complexes are available, it is still difficult to rationally engineer an allosterically regulatable enzyme without decreasing its catalytic activity. Here, we describe an effective strategy to deregulate the allosteric inhibition of enzymes based on the molecular evolution and physicochemical characteristics of allosteric ligand-binding sites. We found that allosteric sites are evolutionarily variable and comprised of more hydrophobic residues than catalytic sites. We applied our findings to design mutations in selected target residues that deregulate the allosteric activity of fructose-1,6-bisphosphatase (FBPase). Specifically, charged amino acids at less conserved positions were substituted with hydrophobic or neutral amino acids with similar sizes. The engineered proteins successfully diminished the allosteric inhibition of E. coli FBPase without affecting its catalytic efficiency. We expect that our method will aid the rational design of enzyme allosteric regulation strategies and facilitate the control of metabolic flux.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Stadtman ER. Allosteric regulation of enzyme activity. Adv Enzymol Relat Areas Mol Biol. 1966;28:41–154. - PubMed
-
- Whitehead E. The regulation of enzyme activity and allosteric transition. Prog Biophys Mol Biol. 1970;21:321–397. - PubMed
-
- Kloosterman H, Hessels GI, Vrijbloed JW, Euverink GJ, Dijkhuizen L. (De)regulation of key enzyme steps in the shikimate pathway and phenylalanine-specific pathway of the actinomycete Amycolatopsis methanolica. Microbiology. 2003;149:3321–3330. - PubMed
-
- Suel GM, Lockless SW, Wall MA, Ranganathan R. Evolutionarily conserved networks of residues mediate allosteric communication in proteins. Nat Struct Biol. 2003;10:59–69. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

