Structural basis for the ABO blood-group dependence of Plasmodium falciparum rosetting
- PMID: 22807674
- PMCID: PMC3395597
- DOI: 10.1371/journal.ppat.1002781
Structural basis for the ABO blood-group dependence of Plasmodium falciparum rosetting
Abstract
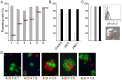
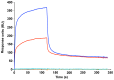
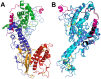
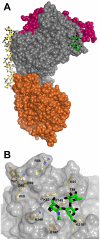
The ABO blood group influences susceptibility to severe Plasmodium falciparum malaria. Recent evidence indicates that the protective effect of group O operates by virtue of reduced rosetting of infected red blood cells (iRBCs) with uninfected RBCs. Rosetting is mediated by a subgroup of PfEMP1 adhesins, with RBC binding being assigned to the N-terminal DBL1α₁ domain. Here, we identify the ABO blood group as the main receptor for VarO rosetting, with a marked preference for group A over group B, which in turn is preferred to group O RBCs. We show that recombinant NTS-DBL1α₁ and NTS-DBL1α₁-CIDR1γ reproduce the VarO-iRBC blood group preference and document direct binding to blood group trisaccharides by surface plasmon resonance. More detailed RBC subgroup analysis showed preferred binding to group A₁, weaker binding to groups A₂ and B, and least binding to groups A(x) and O. The 2.8 Å resolution crystal structure of the PfEMP1-VarO Head region, NTS-DBL1α₁-CIDR1γ, reveals extensive contacts between the DBL1α₁ and CIDR1γ and shows that the NTS-DBL1α₁ hinge region is essential for RBC binding. Computer docking of the blood group trisaccharides and subsequent site-directed mutagenesis localized the RBC-binding site to the face opposite to the heparin-binding site of NTS-DBLα₁. RBC binding involves residues that are conserved between rosette-forming PfEMP1 adhesins, opening novel opportunities for intervention against severe malaria. By deciphering the structural basis of blood group preferences in rosetting, we provide a link between ABO blood grouppolymorphisms and rosette-forming adhesins, consistent with the selective role of falciparum malaria on human genetic makeup.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures




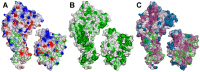
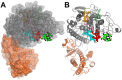
Comment in
-
Revealing the secrets of malaria parasite interaction with blood group A sugars.Pathog Glob Health. 2013 Mar;107(2):45. doi: 10.1179/2047772413Z.000000000113. Pathog Glob Health. 2013. PMID: 23683327 Free PMC article. No abstract available.
References
-
- Boren T, Falk P, Roth KA, Larson G, Normark S. Attachment of Helicobacter pylori to human gastric epithelium mediated by blood group antigens. Science. 1993;262:1892–1895. - PubMed
-
- Hutson AM, Atmar RL, Graham DY, Estes MK. Norwalk virus infection and disease is associated with ABO histo-blood group type. J Infect Dis. 2002;185:1335–1337. - PubMed
-
- Lindesmith L, Moe C, Marionneau S, Ruvoen N, Jiang X, et al. Human susceptibility and resistance to Norwalk virus infection. Nat Med. 2003;9:548–553. - PubMed
-
- Ruiz-Palacios GM, Cervantes LE, Ramos P, Chavez-Munguia B, Newburg DS. Campylobacter jejuni binds intestinal H(O) antigen (Fuc alpha 1, 2Gal beta 1, 4GlcNAc), and fucosyloligosaccharides of human milk inhibit its binding and infection. J Biol Chem. 2003;278:14112–14120. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

