Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n
- PMID: 22807680
- PMCID: PMC3395617
- DOI: 10.1371/journal.ppat.1002800
Endogenous origins of HIV-1 G-to-A hypermutation and restriction in the nonpermissive T cell line CEM2n
Abstract
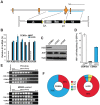
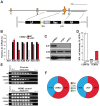
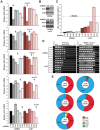
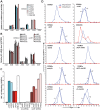
The DNA deaminase APOBEC3G converts cytosines to uracils in retroviral cDNA, which are immortalized as genomic strand G-to-A hypermutations by reverse transcription. A single round of APOBEC3G-dependent mutagenesis can be catastrophic, but evidence suggests that sublethal levels contribute to viral genetic diversity and the associated problems of drug resistance and immune escape. APOBEC3G exhibits an intrinsic preference for the second cytosine in a 5'CC dinucleotide motif leading to 5'GG-to-AG mutations. However, an additional hypermutation signature is commonly observed in proviral sequences from HIV-1 infected patients, 5'GA-to-AA, and it has been attributed controversially to one or more of the six other APOBEC3 deaminases. An unambiguous resolution of this problem has been difficult to achieve, in part due to dominant effects of protein over-expression. Here, we employ gene targeting to dissect the endogenous APOBEC3 contribution to Vif-deficient HIV-1 restriction and hypermutation in a nonpermissive T cell line CEM2n. We report that APOBEC3G-null cells, as predicted from previous studies, lose the capacity to inflict 5'GG-to-AG mutations. In contrast, APOBEC3F-null cells produced viruses with near-normal mutational patterns. Systematic knockdown of other APOBEC3 genes in an APOBEC3F-null background revealed a significant contribution from APOBEC3D in promoting 5'GA-to-AA hypermutations. Furthermore, Vif-deficient HIV-1 restriction was strong in parental CEM2n and APOBEC3D-knockdown cells, partially alleviated in APOBEC3G- or APOBEC3F-null cells, further alleviated in APOBEC3F-null/APOBEC3D-knockdown cells, and alleviated to the greatest extent in APOBEC3F-null/APOBEC3G-knockdown cells revealing clear redundancy in the HIV-1 restriction mechanism. We conclude that endogenous levels of APOBEC3D, APOBEC3F, and APOBEC3G combine to restrict Vif-deficient HIV-1 and cause the hallmark dinucleotide hypermutation patterns in CEM2n. Primary T lymphocytes express a similar set of APOBEC3 genes suggesting that the same repertoire may be important in vivo.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
-
- Malim MH, Emerman M. HIV-1 accessory proteins–ensuring viral survival in a hostile environment. Cell Host Microbe. 2008;3:388–398. - PubMed
-
- Harris RS, Bishop KN, Sheehy AM, Craig HM, Petersen-Mahrt SK, et al. DNA deamination mediates innate immunity to retroviral infection. Cell. 2003;113:803–809. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

