Four loci explain 83% of size variation in the horse
- PMID: 22808074
- PMCID: PMC3394777
- DOI: 10.1371/journal.pone.0039929
Four loci explain 83% of size variation in the horse
Abstract
Horse body size varies greatly due to intense selection within each breed. American Miniatures are less than one meter tall at the withers while Shires and Percherons can exceed two meters. The genetic basis for this variation is not known. We hypothesize that the breed population structure of the horse should simplify efforts to identify genes controlling size. In support of this, here we show with genome-wide association scans (GWAS) that genetic variation at just four loci can explain the great majority of horse size variation. Unlike humans, which are naturally reproducing and possess many genetic variants with weak effects on size, we show that horses, like other domestic mammals, carry just a small number of size loci with alleles of large effect. Furthermore, three of our horse size loci contain the LCORL, HMGA2 and ZFAT genes that have previously been found to control human height. The LCORL/NCAPG locus is also implicated in cattle growth and HMGA2 is associated with dog size. Extreme size diversification is a hallmark of domestication. Our results in the horse, complemented by the prior work in cattle and dog, serve to pinpoint those very few genes that have played major roles in the rapid evolution of size during domestication.
Conflict of interest statement
Figures

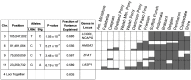
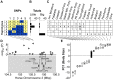
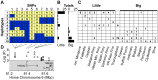
References
-
- Clutton-Brock J, Natural History Museum (London England) A natural history of domesticated mammals. Cambridge, U.K.; New York, NY, USA London;: Cambridge University Press; Natural History Museum. viii, 238 p. p. 1999.
-
- Brooks SA, Makvandi-Nejad S, Chu E, Allen JJ, Streeter C, et al. Morphological variation in the horse: defining complex traits of body size and shape. Animal Genetics. 2010;41:159–165. - PubMed
-
- Gudbjartsson DF, Walters GB, Thorleifsson G, Stefansson H, Halldorsson BV, et al. Many sequence variants affecting diversity of adult human height. Nat Genet. 2008;40:609–615. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

