An improved genetically encoded red fluorescent Ca2+ indicator for detecting optically evoked action potentials
- PMID: 22808076
- PMCID: PMC3393713
- DOI: 10.1371/journal.pone.0039933
An improved genetically encoded red fluorescent Ca2+ indicator for detecting optically evoked action potentials
Abstract
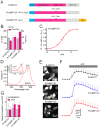
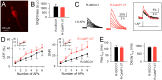
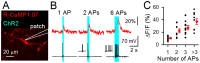
Genetically encoded Ca(2+) indicators (GECIs) are powerful tools to image activities of defined cell populations. Here, we developed an improved red fluorescent GECI, termed R-CaMP1.07, by mutagenizing R-GECO1. In HeLa cell assays, R-CaMP1.07 exhibited a 1.5-2-fold greater fluorescence response compared to R-GECO1. In hippocampal pyramidal neurons, R-CaMP1.07 detected Ca(2+) transients triggered by single action potentials (APs) with a probability of 95% and a signal-to-noise ratio >7 at a frame rate of 50 Hz. The amplitudes of Ca(2+) transients linearly correlated with the number of APs. The expression of R-CaMP1.07 did not significantly alter the electrophysiological properties or synaptic activity patterns. The co-expression of R-CaMP1.07 and channelrhodpsin-2 (ChR2), a photosensitive cation channel, in pyramidal neurons demonstrated that R-CaMP1.07 was applicable for the monitoring of Ca(2+) transients in response to optically evoked APs, because the excitation light for R-CaMP1.07 hardly activated ChR2. These technical advancements provide a novel strategy for monitoring and manipulating neuronal activity with single cell resolution.
Conflict of interest statement
Figures

References
-
- Palmer AE, Tsien RY. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat Protoc. 2006;1:1057–1065. - PubMed
-
- Takahashi N, Oba S, Yukinawa N, Ujita S, Mizunuma M, et al. High-speed multineuron calcium imaging using Nipkow-type confocal microscopy. Curr Protoc Neurosci Chapter 2: Unit 2 14. 2011. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

