Testing the limits of 454 pyrotag sequencing: reproducibility, quantitative assessment and comparison to T-RFLP fingerprinting of aquifer microbes
- PMID: 22808168
- PMCID: PMC3395703
- DOI: 10.1371/journal.pone.0040467
Testing the limits of 454 pyrotag sequencing: reproducibility, quantitative assessment and comparison to T-RFLP fingerprinting of aquifer microbes
Abstract
The characterization of microbial community structure via 16S rRNA gene profiling has been greatly advanced in recent years by the introduction of amplicon pyrosequencing. The possibility of barcoding gives the opportunity to massively screen multiple samples from environmental or clinical sources for community details. However, an on-going debate questions the reproducibility and semi-quantitative rigour of pyrotag sequencing, similar to the early days of community fingerprinting. In this study we demonstrate the reproducibility of bacterial 454 pyrotag sequencing over biological and technical replicates of aquifer sediment bacterial communities. Moreover, we explore the potential of recovering specific template ratios via quantitatively defined template spiking to environmental DNA. We sequenced pyrotag libraries of triplicate sediment samples taken in annual sampling campaigns at a tar oil contaminated aquifer in Düsseldorf, Germany. The abundance of dominating lineages was highly reproducible with a maximal standard deviation of ~4% read abundance across biological, and ~2% across technical replicates. Our workflow also allows for the linking of read abundances within defined assembled pyrotag contigs to that of specific 'in vivo' fingerprinting signatures. Thus we demonstrate that both terminal restriction fragment length polymorphism (T-RFLP) analysis and pyrotag sequencing are capable of recovering highly comparable community structure. Overall diversity was roughly double in amplicon sequencing. Pyrotag libraries were also capable of linearly recovering increasing ratios (up to 20%) of 16S rRNA gene amendments from a pure culture of Aliivibrio fisheri spiked to sediment DNA. Our study demonstrates that 454 pyrotag sequencing is a robust and reproducible method, capable of reliably recovering template abundances and overall community structure within natural microbial communities.
Conflict of interest statement
Figures
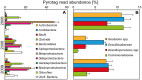
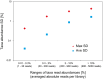

References
-
- Novais RC, Thorstenson YR. The evolution of Pyrosequencing (R) for microbiology: From genes to genomes. J Microbiol Meth. 2011;86:1–7. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

