The role of nuclear matrix proteins binding to matrix attachment regions (Mars) in prostate cancer cell differentiation
- PMID: 22808207
- PMCID: PMC3394767
- DOI: 10.1371/journal.pone.0040617
The role of nuclear matrix proteins binding to matrix attachment regions (Mars) in prostate cancer cell differentiation
Abstract
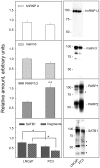
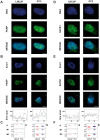
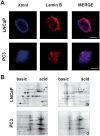
In tumor progression definite alterations in nuclear matrix (NM) protein composition as well as in chromatin structure occur. The NM interacts with chromatin via specialized DNA sequences called matrix attachment regions (MARs). In the present study, using a proteomic approach along with a two-dimensional Southwestern assay and confocal laser microscopy, we show that the differentiation of stabilized human prostate carcinoma cells is marked out by modifications both NM protein composition and bond between NM proteins and MARs. Well-differentiated androgen-responsive and slowly growing LNCaP cells are characterized by a less complex pattern and by a major number of proteins binding MAR sequences in comparison to 22Rv1 cells expressing androgen receptor but androgen-independent. Finally, in the poorly differentiated and strongly aggressive androgen-independent PC3 cells the complexity of NM pattern further increases and a minor number of proteins bind the MARs. Furthermore, in this cell line with respect to LNCaP cells, these changes are synchronous with modifications in both the nuclear distribution of the MAR sequences and in the average loop dimensions that significantly increase. Although the expression of many NM proteins changes during dedifferentiation, only a very limited group of MAR-binding proteins seem to play a key role in this process. Variations in the expression of poly (ADP-ribose) polymerase (PARP) and special AT-rich sequence-binding protein-1 (SATB1) along with an increase in the phosphorylation of lamin B represent changes that might trigger passage towards a more aggressive phenotype. These results suggest that elucidating the MAR-binding proteins that are involved in the differentiation of prostate cancer cells could be an important tool to improve our understanding of this carcinogenesis process, and they could also be novel targets for prostate cancer therapy.
Conflict of interest statement
Figures


References
-
- Lever E, Sheer D. The role of nuclear organization in cancer. J Pathol. 2010;220:114–125. - PubMed
-
- Gluch A, Vidakovic M, Bode J. Scaffold/matrix attachment regions (S/MARs): relevance for disease and therapy. Handb Exp Pharmacol. 2008;186:67–103. - PubMed
-
- Razin SV. The nuclear matrix and chromosomal DNA loops: is their any correlation between partitioning of the genome into loops and functional domains? Cell Mol Biol Lett. 2001;6:59–69. - PubMed
-
- Barboro P, D’Arrigo C, Repaci E, Bagnasco L, Orecchia P, et al. Proteomic analysis of the nuclear matrix in the early stages of rat liver carcinogenesis: identification of differentially expressed and MAR-binding proteins. Exp Cell Res. 2009;315:226–239. - PubMed
-
- Barboro P, D’Arrigo C, Repaci E, Patrone E, Balbi C. Organization of the lamin scaffold in the internal nuclear matrix of normal and transformed hepatocytes. Exp Cell Res. 2010;316:992–1001. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

