The inhibition of anti-DNA binding to DNA by nucleic acid binding polymers
- PMID: 22808279
- PMCID: PMC3394750
- DOI: 10.1371/journal.pone.0040862
The inhibition of anti-DNA binding to DNA by nucleic acid binding polymers
Abstract
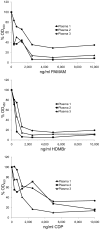
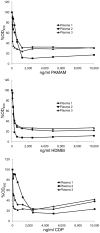
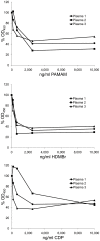
Antibodies to DNA (anti-DNA) are the serological hallmark of systemic lupus erythematosus (SLE) and can mediate disease pathogenesis by the formation of immune complexes. Since blocking immune complex formation can attenuate disease manifestations, the effects of nucleic acid binding polymers (NABPs) on anti-DNA binding in vitro were investigated. The compounds tested included polyamidoamine dendrimer, 1,4-diaminobutane core, generation 3.0 (PAMAM-G3), hexadimethrine bromide, and a β-cylodextrin-containing polycation. As shown with plasma from patients with SLE, NABPs can inhibit anti-DNA antibody binding in ELISA assays. The inhibition was specific since the NABPs did not affect binding to tetanus toxoid or the Sm protein, another lupus autoantigen. Furthermore, the polymers could displace antibody from preformed complexes. Together, these results indicate that NABPs can inhibit the formation of immune complexes and may represent a new approach to treatment.
Conflict of interest statement
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

