Structure-function analysis of Methanobacterium thermoautotrophicum RNA ligase - engineering a thermostable ATP independent enzyme
- PMID: 22809063
- PMCID: PMC3514331
- DOI: 10.1186/1471-2199-13-24
Structure-function analysis of Methanobacterium thermoautotrophicum RNA ligase - engineering a thermostable ATP independent enzyme
Abstract
Background: RNA ligases are essential reagents for many methods in molecular biology including NextGen RNA sequencing. To prevent ligation of RNA to itself, ATP independent mutant ligases, defective in self-adenylation, are often used in combination with activated pre-adenylated linkers. It is important that these ligases not have de-adenylation activity, which can result in activation of RNA and formation of background ligation products. An additional useful feature is for the ligase to be active at elevated temperatures. This has the advantage or reducing preferences caused by structures of single-stranded substrates and linkers.
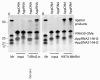
Results: To create an RNA ligase with these desirable properties we performed mutational analysis of the archaeal thermophilic RNA ligase from Methanobacterium thermoautotrophicum. We identified amino acids essential for ATP binding and reactivity but dispensable for phosphodiester bond formation with 5' pre-adenylated donor substrate. The motif V lysine mutant (K246A) showed reduced activity in the first two steps of ligation reaction. The mutant has full ligation activity with pre-adenylated substrates but retained the undesirable activity of deadenylation, which is the reverse of step 2 adenylation. A second mutant, an alanine substitution for the catalytic lysine in motif I (K97A) abolished activity in the first two steps of the ligation reaction, but preserved wild type ligation activity in step 3. The activity of the K97A mutant is similar with either pre-adenylated RNA or single-stranded DNA (ssDNA) as donor substrates but we observed two-fold preference for RNA as an acceptor substrate compared to ssDNA with an identical sequence. In contrast, truncated T4 RNA ligase 2, the commercial enzyme used in these applications, is significantly more active using pre-adenylated RNA as a donor compared to pre-adenylated ssDNA. However, the T4 RNA ligases are ineffective in ligating ssDNA acceptors.
Conclusions: Mutational analysis of the heat stable RNA ligase from Methanobacterium thermoautotrophicum resulted in the creation of an ATP independent ligase. The K97A mutant is defective in the first two steps of ligation but retains full activity in ligation of either RNA or ssDNA to a pre-adenylated linker. The ability of the ligase to function at 65°C should reduce the constraints of RNA secondary structure in RNA ligation experiments.
Figures
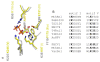


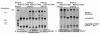

References
-
- Nichols NM, Tabor S, McReynolds LA. RNA ligases. Curr Protoc Mol Biol. 2008;Chapter 3:Unit 3.15. - PubMed
-
- Nandakumar J, Ho CK, Lima CD, Shuman S. RNA substrate specificity and structure-guided mutational analysis of bacteriophage T4 RNA ligase 2. J Biol Chem. 2004;279(30):31337–31347. - PubMed
-
- Blondal T, Thorisdottir A, Unnsteinsdottir U, Hjorleifsdottir S, Aevarsson A, Ernstsson S, Fridjonsson OH, Skirnisdottir S, Wheat JO, Hermannsdottir AG. et al. Isolation and characterization of a thermostable RNA ligase 1 from a Thermus scotoductus bacteriophage TS2126 with good single-stranded DNA ligation properties. Nucleic Acids Res. 2005;33(1):135–142. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

