Derived SNP alleles are used more frequently than ancestral alleles as risk-associated variants in common human diseases
- PMID: 22809343
- PMCID: PMC3655427
- DOI: 10.1142/S0219720012410089
Derived SNP alleles are used more frequently than ancestral alleles as risk-associated variants in common human diseases
Abstract
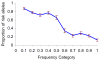
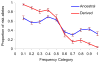
Evolutionary aspects of the genetic architecture of common human diseases remain enigmatic. The results of more than 200 genome-wide association studies published to date were compiled in a catalog (). We used cataloged data to determine whether derived (mutant) alleles are associated with higher risk of human disease more frequently than ancestral alleles. We placed all allelic variants into ten categories of population frequency (0%-100%) in 10% increments. We then analyzed the relationship between allelic frequency, evolutionary status of the polymorphic site (ancestral versus derived), and disease risk status (risk versus protection). Given the same population frequency, derived alleles are more likely to be risk associated than ancestral alleles, as are rarer alleles. The common interpretation of this association is that negative selection prevents fixation of the risk variants. However, disease stratification as early or late onset suggests that weak selection against risk-associated alleles is unlikely a major factor shaping genetic architecture of common diseases. Our results clearly suggest that the duration of existence of an allele in a population is more important. Alleles existing longer tend to show weaker linkage disequilibrium with neighboring alleles, including the causal alleles, and are less likely to tag a SNP-disease association.
Figures


Similar articles
-
Functional and Structural Consequence of Rare Exonic Single Nucleotide Polymorphisms: One Story, Two Tales.Genome Biol Evol. 2015 Oct 9;7(10):2929-40. doi: 10.1093/gbe/evv191. Genome Biol Evol. 2015. PMID: 26454016 Free PMC article.
-
Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases.PLoS Genet. 2015 Jul 22;11(7):e1005371. doi: 10.1371/journal.pgen.1005371. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26201053 Free PMC article.
-
An evolutionary framework for common diseases: the ancestral-susceptibility model.Trends Genet. 2005 Nov;21(11):596-601. doi: 10.1016/j.tig.2005.08.007. Epub 2005 Sep 8. Trends Genet. 2005. PMID: 16153740 Review.
-
Disease-associated alleles in genome-wide association studies are enriched for derived low frequency alleles relative to HapMap and neutral expectations.BMC Med Genomics. 2010 Dec 10;3:57. doi: 10.1186/1755-8794-3-57. BMC Med Genomics. 2010. PMID: 21143973 Free PMC article.
-
On selecting markers for association studies: patterns of linkage disequilibrium between two and three diallelic loci.Genet Epidemiol. 2003 Jan;24(1):57-67. doi: 10.1002/gepi.10217. Genet Epidemiol. 2003. PMID: 12508256 Review.
Cited by
-
Functional and Structural Consequence of Rare Exonic Single Nucleotide Polymorphisms: One Story, Two Tales.Genome Biol Evol. 2015 Oct 9;7(10):2929-40. doi: 10.1093/gbe/evv191. Genome Biol Evol. 2015. PMID: 26454016 Free PMC article.
-
Reproducible simulations of realistic samples for next-generation sequencing studies using Variant Simulation Tools.Genet Epidemiol. 2015 Jan;39(1):45-52. doi: 10.1002/gepi.21867. Epub 2014 Nov 13. Genet Epidemiol. 2015. PMID: 25395236 Free PMC article.
-
Allelic Spectra of Risk SNPs Are Different for Environment/Lifestyle Dependent versus Independent Diseases.PLoS Genet. 2015 Jul 22;11(7):e1005371. doi: 10.1371/journal.pgen.1005371. eCollection 2015 Jul. PLoS Genet. 2015. PMID: 26201053 Free PMC article.
-
Dissecting Generalizability and Actionability of Disease-Associated Genes From 20 Worldwide Ethnolinguistic Cultural Groups.Front Genet. 2022 Jun 24;13:835713. doi: 10.3389/fgene.2022.835713. eCollection 2022. Front Genet. 2022. PMID: 35812734 Free PMC article.
-
Asymmetric Dimethylaminohydrolase Gene Polymorphisms Associated with Preeclampsia Comorbid with HIV Infection in Pregnant Women of African Ancestry.Int J Mol Sci. 2025 Apr 1;26(7):3271. doi: 10.3390/ijms26073271. Int J Mol Sci. 2025. PMID: 40244094 Free PMC article.
References
-
- Morton NE. Into the post-HapMap era. Adv Genet. 2008;60:727–742. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

