A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity
- PMID: 22810204
- PMCID: PMC3488204
- DOI: 10.1093/nar/gks636
A high-throughput, quantitative cell-based screen for efficient tailoring of RNA device activity
Abstract
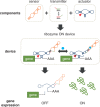
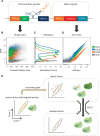
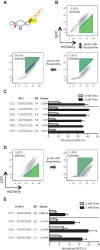
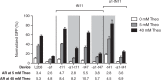
Recent advances have demonstrated the use of RNA-based control devices to program sophisticated cellular functions; however, the efficiency with which these devices can be quantitatively tailored has limited their broader implementation in cellular networks. Here, we developed a high-efficiency, high-throughput and quantitative two-color fluorescence-activated cell sorting-based screening strategy to support the rapid generation of ribozyme-based control devices with user-specified regulatory activities. The high-efficiency of this screening strategy enabled the isolation of a single functional sequence from a library of over 10(6) variants within two sorting cycles. We demonstrated the versatility of our approach by screening large libraries generated from randomizing individual components within the ribozyme device platform to efficiently isolate new device sequences that exhibit increased in vitro cleavage rates up to 10.5-fold and increased in vivo activation ratios up to 2-fold. We also identified a titratable window within which in vitro cleavage rates and in vivo gene-regulatory activities are correlated, supporting the importance of optimizing RNA device activity directly in the cellular environment. Our two-color fluorescence-activated cell sorting-based screen provides a generalizable strategy for quantitatively tailoring genetic control elements for broader integration within biological networks.
Figures



References
-
- Ro D-K, Paradise EM, Ouellet M, Fisher KJ, Newman KL, Ndungu JM, Ho KA, Eachus RA, Ham TS, Kirby J, et al. Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature. 2006;440:940–943. - PubMed
-
- Joyce GF. Forty years of in vitro evolution. Angew. Chem. Int. Ed. Engl. 2007;46:6420–6436. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

