Spread of a distinct Stx2-encoding phage prototype among Escherichia coli O104:H4 strains from outbreaks in Germany, Norway, and Georgia
- PMID: 22811533
- PMCID: PMC3457275
- DOI: 10.1128/JVI.00986-12
Spread of a distinct Stx2-encoding phage prototype among Escherichia coli O104:H4 strains from outbreaks in Germany, Norway, and Georgia
Abstract
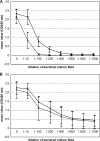
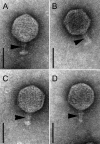
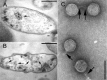
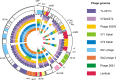
Shiga toxin 2 (Stx2)-producing Escherichia coli (STEC) O104:H4 caused one of the world's largest outbreaks of hemorrhagic colitis and hemolytic uremic syndrome in Germany in 2011. These strains have evolved from enteroaggregative E. coli (EAEC) by the acquisition of the Stx2 genes and have been designated enteroaggregative hemorrhagic E. coli. Nucleotide sequencing has shown that the Stx2 gene is carried by prophages integrated into the chromosome of STEC O104:H4. We studied the properties of Stx2-encoding bacteriophages which are responsible for the emergence of this new type of E. coli pathogen. For this, we analyzed Stx bacteriophages from STEC O104:H4 strains from Germany (in 2001 and 2011), Norway (2006), and the Republic of Georgia (2009). Viable Stx2-encoding bacteriophages could be isolated from all STEC strains except for the Norwegian strain. The Stx2 phages formed lysogens on E. coli K-12 by integration into the wrbA locus, resulting in Stx2 production. The nucleotide sequence of the Stx2 phage P13374 of a German STEC O104:H4 outbreak was determined. From the bioinformatic analyses of the prophage sequence of 60,894 bp, 79 open reading frames were inferred. Interestingly, the Stx2 phages from the German 2001 and 2011 outbreak strains were found to be identical and closely related to the Stx2 phages from the Georgian 2009 isolates. Major proteins of the virion particles were analyzed by mass spectrometry. Stx2 production in STEC O104:H4 strains was inducible by mitomycin C and was compared to Stx2 production of E. coli K-12 lysogens.
Figures




References
-
- Aurass P, Prager R, Flieger A. 2011. EHEC/EAEC O104:H4 strain linked with the 2011 German outbreak of haemolytic uremic syndrome enters into the viable but non-culturable state in response to various stresses and resuscitates upon stress relief. Environ. Microbiol. 13:3139–3148 - PubMed
-
- Baker DR, Moxley RA, Francis DH. 1997. Variation in virulence in the gnotobiotic pig model of O157:H7 Escherichia coli strains of bovine and human origin. Adv. Exp. Med. Biol. 412:53–58 - PubMed
-
- Beutin L, Martin A. 2012. Outbreak of Shiga toxin-producing Escherichia coli (STEC) O104:H4 infection in Germany causes a paradigm shift with regard to human pathogenicity of STEC strains. J. Food Prot. 75:408–418 - PubMed
-
- Beutin L, et al. 2007. Comparative evaluation of the Ridascreen verotoxin enzyme immunoassay for detection of Shiga-toxin producing strains of Escherichia coli (STEC) from food and other sources. J. Appl. Microbiol. 102:630–639 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

