The mechanism of eukaryotic translation initiation: new insights and challenges
- PMID: 22815232
- PMCID: PMC3475172
- DOI: 10.1101/cshperspect.a011544
The mechanism of eukaryotic translation initiation: new insights and challenges
Abstract
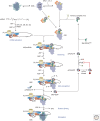
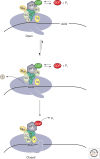
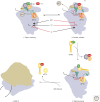
Translation initiation in eukaryotes is a highly regulated and complex stage of gene expression. It requires the action of at least 12 initiation factors, many of which are known to be the targets of regulatory pathways. Here we review our current understanding of the molecular mechanics of eukaryotic translation initiation, focusing on recent breakthroughs from in vitro and in vivo studies. We also identify important unanswered questions that will require new ideas and techniques to solve.
Figures
References
-
- Acker MG, Shin BS, Dever TE, Lorsch JR 2006. Interaction between eukaryotic initiation factors 1A and 5B is required for efficient ribosomal subunit joining. J Biol Chem 281: 8469–8475 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
