Quantifying uniformity of mapped reads
- PMID: 22815359
- PMCID: PMC3467739
- DOI: 10.1093/bioinformatics/bts451
Quantifying uniformity of mapped reads
Abstract
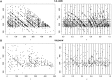
We describe a tool for quantifying the uniformity of mapped reads in high-throughput sequencing experiments. Our statistic directly measures the uniformity of both read position and fragment length, and we explain how to compute a P-value that can be used to quantify biases arising from experimental protocols and mapping procedures. Our method is useful for comparing different protocols in experiments such as RNA-Seq.
Availability and implementation: We provide a freely available and open source python script that can be used to analyze raw read data or reads mapped to transcripts in BAM format at http://www.math.miami.edu/~vhower/ReadSpy.html.
Figures
References
-
- Lander E, Waterman M. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988;2:231–239. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

