Global transcriptome analysis in influenza-infected mouse lungs reveals the kinetics of innate and adaptive host immune responses
- PMID: 22815957
- PMCID: PMC3398930
- DOI: 10.1371/journal.pone.0041169
Global transcriptome analysis in influenza-infected mouse lungs reveals the kinetics of innate and adaptive host immune responses
Abstract
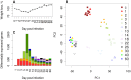
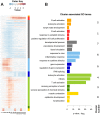
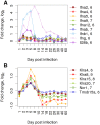
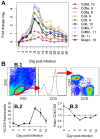
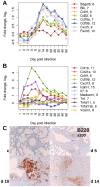
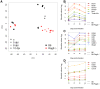
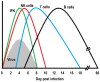
An infection represents a highly dynamic process involving complex biological responses of the host at many levels. To describe such processes at a global level, we recorded gene expression changes in mouse lungs after a non-lethal infection with influenza A virus over a period of 60 days. Global analysis of the large data set identified distinct phases of the host response. The increase in interferon genes and up-regulation of a defined NK-specific gene set revealed the initiation of the early innate immune response phase. Subsequently, infiltration and activation of T and B cells could be observed by an augmentation of T and B cell specific signature gene expression. The changes in B cell gene expression and preceding chemokine subsets were associated with the formation of bronchus-associated lymphoid tissue. In addition, we compared the gene expression profiles from wild type mice with Rag2 mutant mice. This analysis readily demonstrated that the deficiency in the T and B cell responses in Rag2 mutants could be detected by changes in the global gene expression patterns of the whole lung. In conclusion, our comprehensive gene expression study describes for the first time the entire host response and its kinetics to an acute influenza A infection at the transcriptome level.
Conflict of interest statement
Figures
Similar articles
-
Impaired immune responses in the lungs of aged mice following influenza infection.Respir Res. 2009 Nov 18;10(1):112. doi: 10.1186/1465-9921-10-112. Respir Res. 2009. PMID: 19922665 Free PMC article.
-
Initial infectious dose dictates the innate, adaptive, and memory responses to influenza in the respiratory tract.J Leukoc Biol. 2012 Jul;92(1):107-21. doi: 10.1189/jlb.1011490. Epub 2012 Apr 13. J Leukoc Biol. 2012. PMID: 22504848
-
Innate Immunity Induces the Accumulation of Lung Mast Cells During Influenza Infection.Front Immunol. 2018 Oct 4;9:2288. doi: 10.3389/fimmu.2018.02288. eCollection 2018. Front Immunol. 2018. PMID: 30337928 Free PMC article.
-
CD47 Plays a Role as a Negative Regulator in Inducing Protective Immune Responses to Vaccination against Influenza Virus.J Virol. 2016 Jul 11;90(15):6746-6758. doi: 10.1128/JVI.00605-16. Print 2016 Aug 1. J Virol. 2016. PMID: 27194758 Free PMC article.
-
Robust expression of p27Kip1 induced by viral infection is critical for antiviral innate immunity.Cell Microbiol. 2020 Nov;22(11):e13242. doi: 10.1111/cmi.13242. Epub 2020 Aug 1. Cell Microbiol. 2020. PMID: 32596986
Cited by
-
Hematological parameters in the early phase of influenza A virus infection in differentially susceptible inbred mouse strains.BMC Res Notes. 2015 Jun 6;8:225. doi: 10.1186/s13104-015-1195-8. BMC Res Notes. 2015. PMID: 26047817 Free PMC article.
-
System-Based Approaches to Delineate the Antiviral Innate Immune Landscape.Viruses. 2020 Oct 21;12(10):1196. doi: 10.3390/v12101196. Viruses. 2020. PMID: 33096788 Free PMC article. Review.
-
Longitudinal transcriptional analysis of peripheral blood leukocytes in COVID-19 convalescent donors.J Transl Med. 2022 Dec 12;20(1):587. doi: 10.1186/s12967-022-03751-7. J Transl Med. 2022. PMID: 36510222 Free PMC article. Clinical Trial.
-
Infection- and procedure-dependent effects on pulmonary gene expression in the early phase of influenza A virus infection in mice.BMC Microbiol. 2013 Dec 17;13:293. doi: 10.1186/1471-2180-13-293. BMC Microbiol. 2013. PMID: 24341411 Free PMC article.
-
Heterogeneity of Early Host Response to Infection with Four Low-Pathogenic H7 Viruses with a Different Evolutionary History in the Field.Viruses. 2021 Nov 21;13(11):2323. doi: 10.3390/v13112323. Viruses. 2021. PMID: 34835129 Free PMC article.
References
-
- Johnson NP, Mueller J. Updating the accounts: global mortality of the 1918–1920 “Spanish” influenza pandemic. Bull Hist Med. 2002;76:105–115. - PubMed
-
- Fauci AS. Seasonal and pandemic influenza preparedness: science and countermeasures. J Infect Dis. 2006;194:S73–76. - PubMed
-
- Pichlmair A, Reise SC. Innate recognition of viruses. Immunity. 2007;27:370–383. - PubMed
-
- Haller O, Weber F. The interferon response circuit in antiviral host defense. Verh K Acad Geneeskd Belg. 2009;71:73–86. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

