The influence of DNA sequence on epigenome-induced pathologies
- PMID: 22818522
- PMCID: PMC3439399
- DOI: 10.1186/1756-8935-5-11
The influence of DNA sequence on epigenome-induced pathologies
Abstract
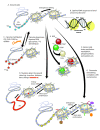
Clear cause-and-effect relationships are commonly established between genotype and the inherited risk of acquiring human and plant diseases and aberrant phenotypes. By contrast, few such cause-and-effect relationships are established linking a chromatin structure (that is, the epitype) with the transgenerational risk of acquiring a disease or abnormal phenotype. It is not entirely clear how epitypes are inherited from parent to offspring as populations evolve, even though epigenetics is proposed to be fundamental to evolution and the likelihood of acquiring many diseases. This article explores the hypothesis that, for transgenerationally inherited chromatin structures, "genotype predisposes epitype", and that epitype functions as a modifier of gene expression within the classical central dogma of molecular biology. Evidence for the causal contribution of genotype to inherited epitypes and epigenetic risk comes primarily from two different kinds of studies discussed herein. The first and direct method of research proceeds by the examination of the transgenerational inheritance of epitype and the penetrance of phenotype among genetically related individuals. The second approach identifies epitypes that are duplicated (as DNA sequences are duplicated) and evolutionarily conserved among repeated patterns in the DNA sequence. The body of this article summarizes particularly robust examples of these studies from humans, mice, Arabidopsis, and other organisms. The bulk of the data from both areas of research support the hypothesis that genotypes predispose the likelihood of displaying various epitypes, but for only a few classes of epitype. This analysis suggests that renewed efforts are needed in identifying polymorphic DNA sequences that determine variable nucleosome positioning and DNA methylation as the primary cause of inherited epigenome-induced pathologies. By contrast, there is very little evidence that DNA sequence directly determines the inherited positioning of numerous and diverse post-translational modifications of histone side chains within nucleosomes. We discuss the medical and scientific implications of these observations on future research and on the development of solutions to epigenetically induced disorders.
Figures
References
-
- Fumagalli M, Cagliani R, Riva S, Pozzoli U, Biasin M, Piacentini L, Comi GP, Bresolin N, Clerici M, Sironi M. Population genetics of IFIH1: ancient population structure, local selection, and implications for susceptibility to type 1 diabetes. Mol Biol Evol. 2010;27:2555–2566. doi: 10.1093/molbev/msq141. - DOI - PubMed
-
- Hayano-Kanashiro C, Calderon-Vazquez C, Ibarra-Laclette E, Herrera-Estrella L, Simpson J. Analysis of gene expression and physiological responses in three Mexican maize landraces under drought stress and recovery irrigation. PLoS One. 2009;4:e7531. doi: 10.1371/journal.pone.0007531. - DOI - PMC - PubMed
-
- Zwonitzer JC, Bubeck DM, Bhattramakki D, Goodman MM, Arellano C, Balint-Kurti PJ. Use of selection with recurrent backcrossing and QTL mapping to identify loci contributing to southern leaf blight resistance in a highly resistant maize line. Theor Appl Genet. 2009;118:911–925. doi: 10.1007/s00122-008-0949-2. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

