Structural and mechanistic basis for the inhibition of Escherichia coli RNA polymerase by T7 Gp2
- PMID: 22819324
- PMCID: PMC3778932
- DOI: 10.1016/j.molcel.2012.06.013
Structural and mechanistic basis for the inhibition of Escherichia coli RNA polymerase by T7 Gp2
Abstract
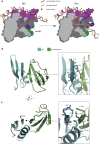
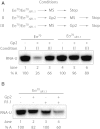
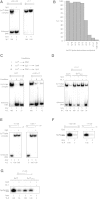
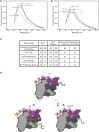
The T7 phage-encoded small protein Gp2 is a non-DNA-binding transcription factor that interacts with the jaw domain of the Escherichia coli (Ec) RNA polymerase (RNAp) β' subunit and inhibits transcriptionally proficient promoter-complex (RPo) formation. Here, we describe the high-resolution solution structure of the Gp2-Ec β' jaw domain complex and show that Gp2 and DNA compete for binding to the β' jaw domain. We reveal that efficient inhibition of RPo formation by Gp2 requires the amino-terminal σ(70) domain region 1.1 (R1.1), and that Gp2 antagonizes the obligatory movement of R1.1 during RPo formation. We demonstrate that Gp2 inhibits RPo formation not just by steric occlusion of the RNAp-DNA interaction but also through long-range antagonistic effects on RNAp-promoter interactions around the RNAp active center that likely occur due to repositioning of R1.1 by Gp2. The inhibition of Ec RNAp by Gp2 thus defines a previously uncharacterized mechanism by which bacterial transcription is regulated by a viral factor.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures


References
-
- Bordes P., Repoila F., Kolb A., Gutierrez C. Involvement of differential efficiency of transcription by esigmas and esigma70 RNA polymerase holoenzymes in growth phase regulation of the Escherichia coli osmE promoter. Mol. Microbiol. 2000;35:845–853. - PubMed
-
- Cámara B., Liu M., Reynolds J., Shadrin A., Liu B., Kwok K., Simpson P., Weinzierl R., Severinov K., Cota E. T7 phage protein Gp2 inhibits the Escherichia coli RNA polymerase by antagonizing stable DNA strand separation near the transcription start site. Proc. Natl. Acad. Sci. USA. 2010;107:2247–2252. - PMC - PubMed
-
- Ederth J., Artsimovitch I., Isaksson L.A., Landick R. The downstream DNA jaw of bacterial RNA polymerase facilitates both transcriptional initiation and pausing. J. Biol. Chem. 2002;277:37456–37463. - PubMed
-
- Ederth J., Mooney R.A., Isaksson L.A., Landick R. Functional interplay between the jaw domain of bacterial RNA polymerase and allele-specific residues in the product RNA-binding pocket. J. Mol. Biol. 2006;356:1163–1179. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources

