CpG island structure and trithorax/polycomb chromatin domains in human cells
- PMID: 22819920
- PMCID: PMC3483364
- DOI: 10.1016/j.ygeno.2012.07.006
CpG island structure and trithorax/polycomb chromatin domains in human cells
Abstract
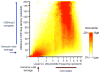
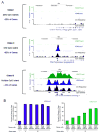
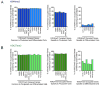
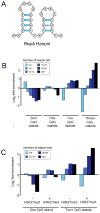
TrxG and PcG complexes play key roles in the epigenetic regulation of development through H3K4me3 and H3K27me3 modification at specific sites throughout the human genome, but how these sites are selected is poorly understood. We find that in pluripotent cells, clustered CpG-islands at genes predict occupancy of H3K4me3 and H3K27me3, and these "bivalent" chromatin domains precisely span the boundaries of CpG-island clusters. These relationships are specific to pluripotent stem cells and are not retained at H3K4me3 and H3K27me3 sites unique to differentiated cells. We show that putative transcripts from clustered CpG-islands predict stem-loop structures characteristic of those bound by PcG complexes, consistent with the possibility that RNA facilitates PcG recruitment or maintenance at these sites. These studies suggest that CpG-island structure plays a fundamental role in establishing developmentally important chromatin structures in the pluripotent genome, and a subordinate role in establishing TrxG/PcG chromatin structure at sites unique to differentiated cells.
Copyright © 2012 Elsevier Inc. All rights reserved.
Figures
Similar articles
-
Genomewide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains.PLoS Genet. 2008 Oct;4(10):e1000242. doi: 10.1371/journal.pgen.1000242. Epub 2008 Oct 31. PLoS Genet. 2008. PMID: 18974828 Free PMC article.
-
Association of the Long Non-coding RNA Steroid Receptor RNA Activator (SRA) with TrxG and PRC2 Complexes.PLoS Genet. 2015 Oct 23;11(10):e1005615. doi: 10.1371/journal.pgen.1005615. eCollection 2015 Oct. PLoS Genet. 2015. PMID: 26496121 Free PMC article.
-
Chromatin remodelling factor Mll1 is essential for neurogenesis from postnatal neural stem cells.Nature. 2009 Mar 26;458(7237):529-33. doi: 10.1038/nature07726. Epub 2009 Feb 11. Nature. 2009. PMID: 19212323 Free PMC article.
-
The interplay of histone modifications - writers that read.EMBO Rep. 2015 Nov;16(11):1467-81. doi: 10.15252/embr.201540945. Epub 2015 Oct 15. EMBO Rep. 2015. PMID: 26474904 Free PMC article. Review.
-
Understanding the interplay between CpG island-associated gene promoters and H3K4 methylation.Biochim Biophys Acta Gene Regul Mech. 2020 Aug;1863(8):194567. doi: 10.1016/j.bbagrm.2020.194567. Epub 2020 Apr 29. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 32360393 Free PMC article. Review.
Cited by
-
PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3.Elife. 2016 May 25;5:e10607. doi: 10.7554/eLife.10607. Elife. 2016. PMID: 27223324 Free PMC article.
-
Discovery of MLL1 binding units, their localization to CpG Islands, and their potential function in mitotic chromatin.BMC Genomics. 2013 Dec 28;14:927. doi: 10.1186/1471-2164-14-927. BMC Genomics. 2013. PMID: 24373511 Free PMC article.
-
Developmentally programmed 3' CpG island methylation confers tissue- and cell-type-specific transcriptional activation.Mol Cell Biol. 2013 May;33(9):1845-58. doi: 10.1128/MCB.01124-12. Epub 2013 Mar 4. Mol Cell Biol. 2013. PMID: 23459939 Free PMC article.
-
A double take on bivalent promoters.Genes Dev. 2013 Jun 15;27(12):1318-38. doi: 10.1101/gad.219626.113. Genes Dev. 2013. PMID: 23788621 Free PMC article. Review.
-
How has the study of the human placenta aided our understanding of partially methylated genes?Epigenomics. 2013 Dec;5(6):645-54. doi: 10.2217/epi.13.62. Epigenomics. 2013. PMID: 24283879 Free PMC article.
References
-
- Ringrose L, Paro R. Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu Rev Genet. 2004;38:413–43. - PubMed
-
- Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128:735–45. - PubMed
-
- Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. - PubMed
-
- Simon JA, Kingston RE. Mechanisms of polycomb gene silencing: knowns and unknowns. Nat Rev Mol Cell Biol. 2009;10:697–708. - PubMed
-
- Surface LE, Thornton SR, Boyer LA. Polycomb group proteins set the stage for early lineage commitment. Cell Stem Cell. 2010;7:288–98. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

