MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis
- PMID: 22820290
- PMCID: PMC3408747
- DOI: 10.1172/JCI63455
MicroRNA-122 plays a critical role in liver homeostasis and hepatocarcinogenesis
Abstract
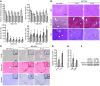
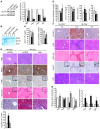
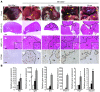
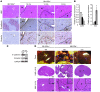
MicroRNA-122 (miR-122), which accounts for 70% of the liver's total miRNAs, plays a pivotal role in the liver. However, its intrinsic physiological roles remain largely undetermined. We demonstrated that mice lacking the gene encoding miR-122a (Mir122a) are viable but develop temporally controlled steatohepatitis, fibrosis, and hepatocellular carcinoma (HCC). These mice exhibited a striking disparity in HCC incidence based on sex, with a male-to-female ratio of 3.9:1, which recapitulates the disease incidence in humans. Impaired expression of microsomal triglyceride transfer protein (MTTP) contributed to steatosis, which was reversed by in vivo restoration of Mttp expression. We found that hepatic fibrosis onset can be partially attributed to the action of a miR-122a target, the Klf6 transcript. In addition, Mir122a(-/-) livers exhibited disruptions in a range of pathways, many of which closely resemble the disruptions found in human HCC. Importantly, the reexpression of miR-122a reduced disease manifestation and tumor incidence in Mir122a(-/-) mice. This study demonstrates that mice with a targeted deletion of the Mir122a gene possess several key phenotypes of human liver diseases, which provides a rationale for the development of a unique therapy for the treatment of chronic liver disease and HCC.
Figures


Comment in
-
miR-122 regulates hepatic lipid metabolism and tumor suppression.J Clin Invest. 2012 Aug;122(8):2773-6. doi: 10.1172/JCI63966. Epub 2012 Jul 23. J Clin Invest. 2012. PMID: 22820284 Free PMC article.
-
miR-122 acts as a tumor suppressor in hepatocarcinogenesis in vivo.J Hepatol. 2013 Apr;58(4):821-3. doi: 10.1016/j.jhep.2012.10.010. Epub 2012 Oct 17. J Hepatol. 2013. PMID: 23085250 No abstract available.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

