Severe acute respiratory syndrome coronavirus accessory proteins 6 and 9b interact in vivo
- PMID: 22820404
- PMCID: PMC7114373
- DOI: 10.1016/j.virusres.2012.07.012
Severe acute respiratory syndrome coronavirus accessory proteins 6 and 9b interact in vivo
Abstract
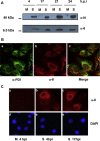
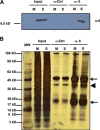
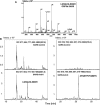
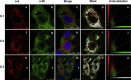
The 3'proximal one-third of the severe acute respiratory syndrome coronavirus (SARS-CoV) genome encodes the structural proteins and eight accessory proteins, including 3a, 3b, 6, 7a, 7b, 8a, 8b and 9b, varying in length from 39 to 274aa which do not share significant homology with viral proteins of known coronaviruses. The SARS-CoV protein 6 is 63 amino acids in length and has been previously involved in virus pathogenicity and replication. To further analyze this functions, the interaction of SARS-CoV protein 6 with other viral and/or cellular factors has been analyzed during SARS-CoV infective cycle. Protein 6 immunoprecipitation from extracts of SARS-CoV infected cells and mass spectrometry analysis revealed an interaction of viral proteins 6 and 9b in biologically relevant conditions. This interaction has been reinforced by co-localization of both proteins in the cytoplasm of SARS-CoV infected cells.
Copyright © 2012 Elsevier B.V. All rights reserved.
Figures
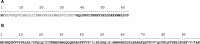
Similar articles
-
Accessory proteins of SARS-CoV and other coronaviruses.Antiviral Res. 2014 Sep;109:97-109. doi: 10.1016/j.antiviral.2014.06.013. Epub 2014 Jul 1. Antiviral Res. 2014. PMID: 24995382 Free PMC article. Review.
-
The human severe acute respiratory syndrome coronavirus (SARS-CoV) 8b protein is distinct from its counterpart in animal SARS-CoV and down-regulates the expression of the envelope protein in infected cells.Virology. 2006 Oct 10;354(1):132-42. doi: 10.1016/j.virol.2006.06.026. Epub 2006 Jul 31. Virology. 2006. PMID: 16876844 Free PMC article.
-
The nonstructural protein 8 (nsp8) of the SARS coronavirus interacts with its ORF6 accessory protein.Virology. 2007 Sep 30;366(2):293-303. doi: 10.1016/j.virol.2007.04.029. Epub 2007 May 25. Virology. 2007. PMID: 17532020 Free PMC article.
-
Inhibition of SARS-CoV replication cycle by small interference RNAs silencing specific SARS proteins, 7a/7b, 3a/3b and S.Antiviral Res. 2007 Mar;73(3):219-27. doi: 10.1016/j.antiviral.2006.10.008. Epub 2006 Nov 7. Antiviral Res. 2007. PMID: 17112601 Free PMC article.
-
Understanding the accessory viral proteins unique to the severe acute respiratory syndrome (SARS) coronavirus.Antiviral Res. 2006 Nov;72(2):78-88. doi: 10.1016/j.antiviral.2006.05.010. Epub 2006 Jun 6. Antiviral Res. 2006. PMID: 16820226 Free PMC article. Review.
Cited by
-
Structure and Function of Major SARS-CoV-2 and SARS-CoV Proteins.Bioinform Biol Insights. 2021 Jun 22;15:11779322211025876. doi: 10.1177/11779322211025876. eCollection 2021. Bioinform Biol Insights. 2021. PMID: 34220199 Free PMC article. Review.
-
Interactions among SARS-CoV accessory proteins revealed by bimolecular fluorescence complementation assay.Acta Pharm Sin B. 2015 Sep;5(5):487-92. doi: 10.1016/j.apsb.2015.05.002. Epub 2015 Jun 6. Acta Pharm Sin B. 2015. PMID: 26579480 Free PMC article.
-
SARS-CoV-2: Insights into its structural intricacies and functional aspects for drug and vaccine development.Int J Biol Macromol. 2021 May 15;179:45-60. doi: 10.1016/j.ijbiomac.2021.02.212. Epub 2021 Mar 1. Int J Biol Macromol. 2021. PMID: 33662418 Free PMC article. Review.
-
The role of severe acute respiratory syndrome (SARS)-coronavirus accessory proteins in virus pathogenesis.Viruses. 2012 Nov 7;4(11):2902-23. doi: 10.3390/v4112902. Viruses. 2012. PMID: 23202509 Free PMC article. Review.
-
Accessory proteins of SARS-CoV and other coronaviruses.Antiviral Res. 2014 Sep;109:97-109. doi: 10.1016/j.antiviral.2014.06.013. Epub 2014 Jul 1. Antiviral Res. 2014. PMID: 24995382 Free PMC article. Review.
References
-
- Calvo E., Escors D., Lopez J.A., Gonzalez J.M., Alvarez A., Arza E., Enjuanes L. Phosphorylation and subcellular localization of transmissible gastroenteritis virus nucleocapsid protein in infected cells. Journal of General Virology. 2005;86(Pt 8):2255–2267. - PubMed
-
- Chan W.S., Wu C., Chow S.C., Cheung T., To K.F., Leung W.K., Chan P.K., Lee K.C., Ng H.K., Au D.M., Lo A.W. Coronaviral hypothetical and structural proteins were found in the intestinal surface enterocytes and pneumocytes of severe acute respiratory syndrome (SARS) Modern Pathology. 2005;18(11):1432–1439. - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

