Fast and accurate genotype imputation in genome-wide association studies through pre-phasing
- PMID: 22820512
- PMCID: PMC3696580
- DOI: 10.1038/ng.2354
Fast and accurate genotype imputation in genome-wide association studies through pre-phasing
Abstract
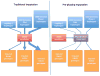
The 1000 Genomes Project and disease-specific sequencing efforts are producing large collections of haplotypes that can be used as reference panels for genotype imputation in genome-wide association studies (GWAS). However, imputing from large reference panels with existing methods imposes a high computational burden. We introduce a strategy called 'pre-phasing' that maintains the accuracy of leading methods while reducing computational costs. We first statistically estimate the haplotypes for each individual within the GWAS sample (pre-phasing) and then impute missing genotypes into these estimated haplotypes. This reduces the computational cost because (i) the GWAS samples must be phased only once, whereas standard methods would implicitly repeat phasing with each reference panel update, and (ii) it is much faster to match a phased GWAS haplotype to one reference haplotype than to match two unphased GWAS genotypes to a pair of reference haplotypes. We implemented our approach in the MaCH and IMPUTE2 frameworks, and we tested it on data sets from the Wellcome Trust Case Control Consortium 2 (WTCCC2), the Genetic Association Information Network (GAIN), the Women's Health Initiative (WHI) and the 1000 Genomes Project. This strategy will be particularly valuable for repeated imputation as reference panels evolve.
Figures
References
Publication types
MeSH terms
Grants and funding
- G1001799/MRC_/Medical Research Council/United Kingdom
- R01 MH084698/MH/NIMH NIH HHS/United States
- R01 HG002651/HG/NHGRI NIH HHS/United States
- DK0855840/DK/NIDDK NIH HHS/United States
- 090532/WT_/Wellcome Trust/United Kingdom
- HG005552/HG/NHGRI NIH HHS/United States
- HGO2585/PHS HHS/United States
- G0801823/MRC_/Medical Research Council/United Kingdom
- RC2 HG005552/HG/NHGRI NIH HHS/United States
- U01 HG006513/HG/NHGRI NIH HHS/United States
- R01 HG007022/HG/NHGRI NIH HHS/United States
- HG005581/HG/NHGRI NIH HHS/United States
- R01 HL117626/HL/NHLBI NIH HHS/United States
- RC2 HG005581/HG/NHGRI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

