Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F
- PMID: 22820988
- PMCID: PMC3414675
- DOI: 10.1038/nsmb.2348
Crystal structures of the JAK2 pseudokinase domain and the pathogenic mutant V617F
Abstract
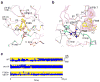
The protein tyrosine kinase JAK2 mediates signaling through numerous cytokine receptors. JAK2 possesses a pseudokinase domain (JH2) and a tyrosine kinase domain (JH1). Through unknown mechanisms, JH2 regulates the catalytic activity of JH1, and hyperactivating mutations in the JH2 region of human JAK2 cause myeloproliferative neoplasms (MPNs). We showed previously that JAK2 JH2 is, in fact, catalytically active. Here we present crystal structures of human JAK2 JH2, including both wild type and the most prevalent MPN mutant, V617F. The structures reveal that JH2 adopts the fold of a prototypical protein kinase but binds Mg-ATP noncanonically. The structural and biochemical data indicate that the V617F mutation rigidifies α-helix C in the N lobe of JH2, facilitating trans-phosphorylation of JH1. The crystal structures of JH2 afford new opportunities for the design of novel JAK2 therapeutics targeting MPNs.
Figures
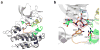
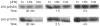

References
-
- Levy DE, Darnell JE., Jr Stats: transcriptional control and biological impact. Nat Rev Mol Cell Biol. 2002;3:651–662. - PubMed
-
- Lucet IS, et al. The structural basis of Janus kinase 2 inhibition by a potent and specific pan-Janus kinase inhibitor. Blood. 2006;107:176–183. - PubMed
-
- Williams NK, et al. Dissecting specificity in the Janus kinases: the structures of JAK-specific inhibitors complexed to the JAK1 and JAK2 protein tyrosine kinase domains. J Mol Biol. 2009;387:219–232. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

