Evolution of viral proteins originated de novo by overprinting
- PMID: 22821011
- PMCID: PMC3494269
- DOI: 10.1093/molbev/mss179
Evolution of viral proteins originated de novo by overprinting
Abstract
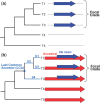
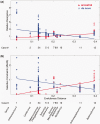
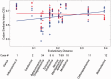
New protein-coding genes can originate either through modification of existing genes or de novo. Recently, the importance of de novo origination has been recognized in eukaryotes, although eukaryotic genes originated de novo are relatively rare and difficult to identify. In contrast, viruses contain many de novo genes, namely those in which an existing gene has been "overprinted" by a new open reading frame, a process that generates a new protein-coding gene overlapping the ancestral gene. We analyzed the evolution of 12 experimentally validated viral genes that originated de novo and estimated their relative ages. We found that young de novo genes have a different codon usage from the rest of the genome. They evolve rapidly and are under positive or weak purifying selection. Thus, young de novo genes might have strain-specific functions, or no function, and would be difficult to detect using current genome annotation methods that rely on the sequence signature of purifying selection. In contrast to young de novo genes, older de novo genes have a codon usage that is similar to the rest of the genome. They evolve slowly and are under stronger purifying selection. Some of the oldest de novo genes evolve under stronger selection pressure than the ancestral gene they overlap, suggesting an evolutionary tug of war between the ancestral and the de novo gene.
Figures

References
-
- Abroi A, Gough J. Are viruses a source of new protein folds for organisms?—virosphere structure space and evolution. Bioessays. 2011;33:626–635. - PubMed
-
- Anisimova M, Kosiol C. Investigating protein-coding sequence evolution with probabilistic codon substitution models. Mol Biol Evol. 2009;26:255–271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

