Proximity of H2A.Z containing nucleosome to the transcription start site influences gene expression levels in the mammalian liver and brain
- PMID: 22821566
- PMCID: PMC3467062
- DOI: 10.1093/nar/gks665
Proximity of H2A.Z containing nucleosome to the transcription start site influences gene expression levels in the mammalian liver and brain
Abstract
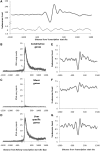
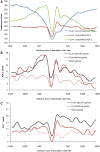
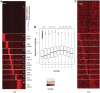
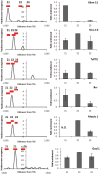
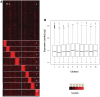
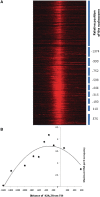
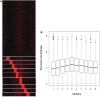
Nucleosome positioning maps of several organisms have shown that Transcription Start Sites (TSSs) are marked by nucleosome depleted regions flanked by strongly positioned nucleosomes. Using genome-wide nucleosome maps and histone variant occupancy in the mouse liver, we show that the majority of genes were associated with a single prominent H2A.Z containing nucleosome in their promoter region. We classified genes into clusters depending on the proximity of H2A.Z to the TSS. The genes with no detectable H2A.Z showed lowest expression level, whereas H2A.Z was positioned closer to the TSS of genes with higher expression levels. We confirmed this relation between the proximity of H2A.Z and expression level in the brain. The proximity of histone variant H2A.Z, but not H3.3 to the TSS, over seven consecutive nucleosomes, was correlated with expression. Further, a nucleosome was positioned over the TSS of silenced genes while it was displaced to expose the TSS in highly expressed genes. Our results suggest that gene expression levels in vivo are determined by accessibility of the TSS and proximity of H2A.Z.
Figures


References
-
- Lantermann AB, Straub T, Stralfors A, Yuan GC, Ekwall K, Korber P. Schizosaccharomyces pombe genome-wide nucleosome mapping reveals positioning mechanisms distinct from those of Saccharomyces cerevisiae. Nat. Struct. Mol. Biol. 2010;17:251–257. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

