Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation
- PMID: 22821944
- PMCID: PMC3468538
- DOI: 10.1152/ajpgi.00152.2012
Colonic mucosal microbiome differs from stool microbiome in cirrhosis and hepatic encephalopathy and is linked to cognition and inflammation
Abstract
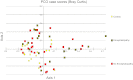
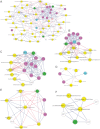
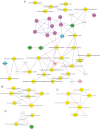
Although hepatic encephalopathy (HE) is linked to the gut microbiota, stool microbiome analysis has not found differences between HE and no-HE patients. This study aimed to compare sigmoid mucosal microbiome of cirrhotic patients to controls, between HE vs. no-HE patients, and to study their linkage with cognition and inflammation. Sixty cirrhotic patients (36 HE and 24 no-HE) underwent cognitive testing, stool collection, cytokine (Th1, Th2, Th17, and innate immunity), and endotoxin analysis. Thirty-six patients (19 HE and 17 no-HE) and 17 age-matched controls underwent sigmoid biopsies. Multitag pyrosequencing (including autochthonous genera, i.e., Blautia, Roseburia, Fecalibacterium, Dorea) was performed on stool and mucosa. Stool and mucosal microbiome differences within/between groups and correlation network analyses were performed. Controls had significantly higher autochthonous and lower pathogenic genera compared with cirrhotic patients, especially HE patients. HE patients had worse MELD (model for end-stage liver disease) score and cognition and higher IL-6 and endotoxin than no-HE. Mucosal microbiota was different from stool within both HE/no-HE groups. Between HE/no-HE patients, there was no difference in stool microbiota but mucosal microbiome was different with lower Roseburia and higher Enterococcus, Veillonella, Megasphaera, and Burkholderia abundance in HE. On network analysis, autochthonous genera (Blautia, Fecalibacterium, Roseburia, and Dorea) were associated with good cognition and decreased inflammation in both HE/no-HE, whereas genera overrepresented in HE (Enterococcus, Megasphaera, and Burkholderia) were linked to poor cognition and inflammation. Sigmoid mucosal microbiome differs significantly from stool microbiome in cirrhosis. Cirrhotic, especially HE, patients' mucosal microbiota is significantly different from controls with a lack of potentially beneficial autochthonous and overgrowth of potentially pathogenic genera, which are associated with poor cognition and inflammation.
Figures
References
-
- Bajaj JS, Gillevet PM, Patel NR, Ahluwalia V, Ridlon JM, Kettenmann B, Schubert CM, Sikaroodi M, Heuman DM, Crossey MME, Bell DE, Hylemon PB, Fatouros PP, Taylor-Robinson SD. A longitudinal systems biology analysis of lactulose withdrawal in hepatic encephalopathy. Metab Brain Dis 27: 205–215, 2012 - PubMed
-
- Bajaj JS, Hafeezullah M, Franco J, Varma RR, Hoffmann RG, Knox JF, Hischke D, Hammeke TA, Pinkerton SD, Saeian K. Inhibitory control test for the diagnosis of minimal hepatic encephalopathy. Gastroenterology 135: 1591–1600, 2008 - PubMed
-
- Bajaj JS, Heuman DM, Wade JB, Gibson DP, Saeian K, Wegelin JA, Hafeezullah M, Bell DE, Sterling RK, Stravitz RT, Fuchs M, Luketic V, Sanyal AJ. Rifaximin improves driving simulator performance in a randomized trial of patients with minimal hepatic encephalopathy. Gastroenterology 140: 478–487, 2011 - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

