The primosomal protein DnaD inhibits cooperative DNA binding by the replication initiator DnaA in Bacillus subtilis
- PMID: 22821970
- PMCID: PMC3430336
- DOI: 10.1128/JB.00958-12
The primosomal protein DnaD inhibits cooperative DNA binding by the replication initiator DnaA in Bacillus subtilis
Abstract
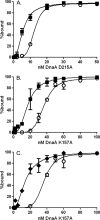
DnaA is an AAA+ ATPase and the conserved replication initiator in bacteria. Bacteria control the timing of replication initiation by regulating the activity of DnaA. DnaA binds to multiple sites in the origin of replication (oriC) and is required for recruitment of proteins needed to load the replicative helicase. DnaA also binds to other chromosomal regions and functions as a transcription factor at some of these sites. Bacillus subtilis DnaD is needed during replication initiation for assembly of the replicative helicase at oriC and during replication restart at stalled replication forks. DnaD associates with DnaA at oriC and at other chromosomal regions bound by DnaA. Using purified proteins, we found that DnaD inhibited the ability of DnaA to bind cooperatively to DNA and caused a decrease in the apparent dissociation constant. These effects of DnaD were independent of the ability of DnaA to bind or hydrolyze ATP. Other proteins known to regulate B. subtilis DnaA also affect DNA binding, whereas much of the regulation of Escherichia coli DnaA affects nucleotide hydrolysis or exchange. We found that the rate of nucleotide exchange for B. subtilis DnaA was high and not affected by DnaD. The rapid exchange is similar to that of Staphylococcus aureus DnaA and in contrast to the low exchange rate of Escherichia coli DnaA. We suggest that organisms in which DnaA has a high rate of nucleotide exchange predominantly regulate the DNA binding activity of DnaA and that those with low rates of exchange regulate hydrolysis and exchange.
Figures



References
-
- Atlung T, Clausen ES, Hansen FG. 1985. Autoregulation of the dnaA gene of Escherichia coli K12. Mol. Gen. Genet. 200:442–450 - PubMed
-
- Bramhill D, Kornberg A. 1988. Duplex opening by dnaA protein at novel sequences in initiation of replication at the origin of the E. coli chromosome. Cell 52:743–755 - PubMed
-
- Braun RE, O'Day K, Wright A. 1985. Autoregulation of the DNA replication gene dnaA in E. coli K-12. Cell 40:159–169 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

