Two ATPases
- PMID: 22822068
- PMCID: PMC3436262
- DOI: 10.1074/jbc.X112.402313
Two ATPases
Abstract
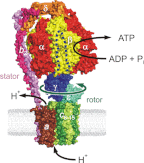
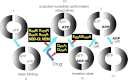
In this article, I reflect on research on two ATPases. The first is F(1)F(0)-ATPase, also known as ATP synthase. It is the terminal enzyme in oxidative phosphorylation and famous as a nanomotor. Early work on mitochondrial enzyme involved purification in large amount, followed by deduction of subunit composition and stoichiometry and determination of molecular sizes of holoenzyme and individual subunits. Later work on Escherichia coli enzyme utilized mutagenesis and optical probes to reveal the molecular mechanism of ATP hydrolysis and detailed facets of catalysis. The second ATPase is P-glycoprotein, which confers multidrug resistance, notably to anticancer drugs, in mammalian cells. Purification of the protein in large quantity allowed detailed characterization of catalysis, formulation of an alternating sites mechanism, and recently, advances in structural characterization.
Figures






Similar articles
-
Rotational catalysis in proton pumping ATPases: from E. coli F-ATPase to mammalian V-ATPase.Biochim Biophys Acta. 2012 Oct;1817(10):1711-21. doi: 10.1016/j.bbabio.2012.03.015. Epub 2012 Mar 20. Biochim Biophys Acta. 2012. PMID: 22459334 Review.
-
Amino Acid Residues β139, β189, and β319 Modulate ADP-Inhibition in Escherichia coli H+-FOF1-ATP Synthase.Biochemistry (Mosc). 2019 Apr;84(4):407-415. doi: 10.1134/S0006297919040084. Biochemistry (Mosc). 2019. PMID: 31228932
-
The role of the epsilon subunit in the Escherichia coli ATP synthase. The C-terminal domain is required for efficient energy coupling.J Biol Chem. 2006 Jan 6;281(1):501-7. doi: 10.1074/jbc.M509986200. Epub 2005 Nov 2. J Biol Chem. 2006. PMID: 16267041
-
Inhibition of ATP hydrolysis by thermoalkaliphilic F1Fo-ATP synthase is controlled by the C terminus of the epsilon subunit.J Bacteriol. 2006 Jun;188(11):3796-804. doi: 10.1128/JB.00040-06. J Bacteriol. 2006. PMID: 16707672 Free PMC article.
-
Rotating proton pumping ATPases: subunit/subunit interactions and thermodynamics.IUBMB Life. 2013 Mar;65(3):247-54. doi: 10.1002/iub.1134. IUBMB Life. 2013. PMID: 23441040 Review.
Cited by
-
The strong in vivo anti-tumor effect of the UIC2 monoclonal antibody is the combined result of Pgp inhibition and antibody dependent cell-mediated cytotoxicity.PLoS One. 2014 Sep 19;9(9):e107875. doi: 10.1371/journal.pone.0107875. eCollection 2014. PLoS One. 2014. PMID: 25238617 Free PMC article.
-
Functional importance of αIle-346 and αIle-348 in the catalytic sites of Escherichia coli ATP synthase.Arch Biochem Biophys. 2016 Feb 15;592:27-37. doi: 10.1016/j.abb.2016.01.009. Epub 2016 Jan 14. Arch Biochem Biophys. 2016. PMID: 26775572 Free PMC article.
-
Revealing the fate of cell surface human P-glycoprotein (ABCB1): The lysosomal degradation pathway.Biochim Biophys Acta. 2015 Oct;1853(10 Pt A):2361-70. doi: 10.1016/j.bbamcr.2015.06.001. Epub 2015 Jun 6. Biochim Biophys Acta. 2015. PMID: 26057472 Free PMC article.
-
Advances of medical nanorobots for future cancer treatments.J Hematol Oncol. 2023 Jul 14;16(1):74. doi: 10.1186/s13045-023-01463-z. J Hematol Oncol. 2023. PMID: 37452423 Free PMC article. Review.
-
Substrates Control Multimerization and Activation of the Multi-Domain ATPase Motor of Type VII Secretion.Cell. 2015 Apr 23;161(3):501-512. doi: 10.1016/j.cell.2015.03.040. Epub 2015 Apr 9. Cell. 2015. PMID: 25865481 Free PMC article.
References
-
- Sherratt H. S. A. (1986) Hypoglycin, the famous toxin of the unripe Jamaican ackee fruit. Trends Pharmacol. Sci. 7, 186–191
-
- Schulz H., Fong J. C. (1981) 4-Pentenoic acid. Methods Enzymol. 72, 604–610 - PubMed
-
- Senior A. E., MacLennan D. H. (1970) Mitochondrial “structural protein.” A reassessment. J. Biol. Chem. 245, 5086–5095 - PubMed
-
- Senior A. E., Brooks J. C. (1970) Studies on the mitochondrial oligomycin-insensitive ATPase. I. An improved method of purification and the behavior of the enzyme in solutions of various depolymerizing agents. Arch. Biochem. Biophys. 140, 257–266 - PubMed
-
- Senior A. E., Brooks J. C. (1971) The subunit composition of the mitochondrial oligomycin-insensitive ATPase. FEBS Lett. 17, 327–329 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

