Multiple independent analyses reveal only transcription factors as an enriched functional class associated with microRNAs
- PMID: 22824421
- PMCID: PMC3430561
- DOI: 10.1186/1752-0509-6-90
Multiple independent analyses reveal only transcription factors as an enriched functional class associated with microRNAs
Abstract
Background: Transcription factors (TFs) have long been known to be principally activators of transcription in eukaryotes and prokaryotes. The growing awareness of the ubiquity of microRNAs (miRNAs) as suppressive regulators in eukaryotes, suggests the possibility of a mutual, preferential, self-regulatory connectivity between miRNAs and TFs. Here we investigate the connectivity from TFs and miRNAs to other genes and each other using text mining, TF promoter binding site and 6 different miRNA binding site prediction methods.
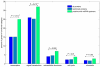
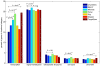
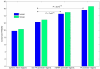
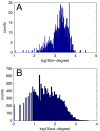
Results: In the first approach text mining of PubMed abstracts reveal statistically significant associations between miRNAs and both TFs and signal transduction gene classes. Secondly, prediction of miRNA targets in human and mouse 3'UTRs show enrichment only for TFs but not consistently across prediction methods for signal transduction or other gene classes. Furthermore, a random sample of 986 TarBase entries was scored for experimental evidence by manual inspection of the original papers, and enrichment for TFs was observed to increase with score. Low-scoring TarBase entries, where experimental evidence is anticorrelated miRNA:mRNA expression with predicted miRNA targets, appear not to select for real miRNA targets to any degree. Our manually validated text-mining results also suggests that miRNAs may be activated by more TFs than other classes of genes, as 7% of miRNA:TF co-occurrences in the literature were TFs activating miRNAs. This was confirmed when thirdly, we found enrichment for predicted, conserved TF binding sites in miRNA and TF genes compared to other gene classes.
Conclusions: We see enrichment of connections between miRNAs and TFs using several independent methods, suggestive of a network of mutual activating and suppressive regulation. We have also built regulatory networks (containing 2- and 3-loop motifs) for mouse and human using predicted miRNA and TF binding sites and we have developed a web server to search and display these loops, available for the community at http://rth.dk/resources/tfmirloop.
Figures

References
-
- Chen K, Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat Rev Genet. 2007;8(2):93–103. - PubMed
-
- Cobb BS, Nesterova TB, Thompson E, Hertweck A, O'Connor E, Godwin J, Wilson CB, Brockdorff N, Fisher AG, Smale ST, Merkenschlager M. T cell lineage choice and differentiation in the absence of the RNase III enzyme Dicer. J Exp Med. 2005;201(9):1367–1373. doi: 10.1084/jem.20050572. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

