Chromosomes without a 30-nm chromatin fiber
- PMID: 22825571
- PMCID: PMC3474659
- DOI: 10.4161/nucl.21222
Chromosomes without a 30-nm chromatin fiber
Abstract
How is a long strand of genomic DNA packaged into a mitotic chromosome or nucleus? The nucleosome fiber (beads-on-a-string), in which DNA is wrapped around core histones, has long been assumed to be folded into a 30-nm chromatin fiber, and a further helically folded larger fiber. However, when frozen hydrated human mitotic cells were observed using cryoelectron microscopy, no higher-order structures that included 30-nm chromatin fibers were found. To investigate the bulk structure of mitotic chromosomes further, we performed small-angle X-ray scattering (SAXS), which can detect periodic structures in noncrystalline materials in solution. The results were striking: no structural feature larger than 11 nm was detected, even at a chromosome-diameter scale (~1 μm). We also found a similar scattering pattern in interphase nuclei of HeLa cells in the range up to ~275 nm. Our findings suggest a common structural feature in interphase and mitotic chromatins: compact and irregular folding of nucleosome fibers occurs without a 30-nm chromatin structure.
Figures

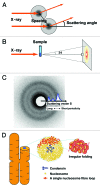
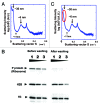
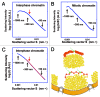
Comment on
- Nishino Y, Eltsov M, Joti Y, Ito K, Takata H, Takahashi Y, et al. Human mitotic chromosomes consist predominantly of irregularly folded nucleosome fibres without a 30-nm chromatin structure. EMBO J. 2012;31:1644–53. doi: 10.1038/emboj.2012.35.
References
-
- Alberts B, Johnson A, Lewis J, Raff M, Roberts K, Walter P. Molecular Biology of the Cell, Fifth Edition 2007.
-
- Olins DE, Olins AL. Chromatin history: our view from the bridge. Nat Rev Mol Cell Biol. 2003;4:809–14. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
