DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation
- PMID: 22825850
- PMCID: PMC3436322
- DOI: 10.1074/jbc.M112.390120
DNA damage-induced heterogeneous nuclear ribonucleoprotein K sumoylation regulates p53 transcriptional activation
Abstract
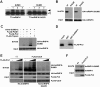
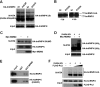
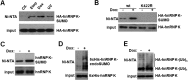
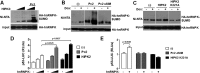
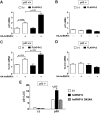
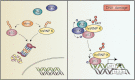
Heterogeneous nuclear ribonucleoprotein (hnRNP) K is a nucleocytoplasmic shuttling protein that is a key player in the p53-triggered DNA damage response, acting as a cofactor for p53 in response to DNA damage. hnRNP K is a substrate of the ubiquitin E3 ligase MDM2 and, upon DNA damage, is de-ubiquitylated. In sharp contrast with the role and consequences of the other post-translational modifications, nothing is known about the role of SUMO conjugation to hnRNP K in p53 transcriptional co-activation. In the present work, we show that hnRNP K is modified by SUMO in lysine 422 within its KH3 domain, and sumoylation is regulated by the E3 ligase Pc2/CBX4. Most interestingly, DNA damage stimulates hnRNP K sumoylation through Pc2 E3 activity, and this modification is required for p53 transcriptional activation. Abrogation of hnRNP K sumoylation leads to an aberrant regulation of the p53 target gene p21. Our findings link the DNA damage-induced Pc2 activation to the p53 transcriptional co-activation through hnRNP K sumoylation.
Figures


References
-
- Geiss-Friedlander R., Melchior F. (2007) Concepts in sumoylation: a decade on. Nat. Rev. Mol. Cell Biol. 8, 947–956 - PubMed
-
- Hay R. T. (2005) SUMO: a history of modification. Mol. Cell 18, 1–12 - PubMed
-
- Bernassola F., Karin M., Ciechanover A., Melino G. (2008) The HECT family of E3 ubiquitin ligases: multiple players in cancer development. Cancer Cell 14, 10–21 - PubMed
-
- Deshaies R. J., Joazeiro C. A. (2009) RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78, 399–434 - PubMed
-
- Kahyo T., Nishida T., Yasuda H. (2001) Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 8, 713–718 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

