Atg9 vesicles are an important membrane source during early steps of autophagosome formation
- PMID: 22826123
- PMCID: PMC3410421
- DOI: 10.1083/jcb.201202061
Atg9 vesicles are an important membrane source during early steps of autophagosome formation
Abstract
During the process of autophagy, cytoplasmic materials are sequestered by double-membrane structures, the autophagosomes, and then transported to a lytic compartment to be degraded. One of the most fundamental questions about autophagy involves the origin of the autophagosomal membranes. In this study, we focus on the intracellular dynamics of Atg9, a multispanning membrane protein essential for autophagosome formation in yeast. We found that the vast majority of Atg9 existed on cytoplasmic mobile vesicles (designated Atg9 vesicles) that were derived from the Golgi apparatus in a process involving Atg23 and Atg27. We also found that only a few Atg9 vesicles were required for a single round of autophagosome formation. During starvation, several Atg9 vesicles assembled individually into the preautophagosomal structure, and eventually, they are incorporated into the autophagosomal outer membrane. Our findings provide conclusive linkage between the cytoplasmic Atg9 vesicles and autophagosomal membranes and offer new insight into the requirement for Atg9 vesicles at the early step of autophagosome formation.
Figures
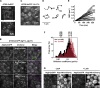
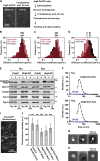
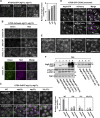
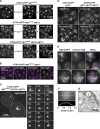
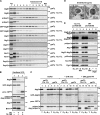

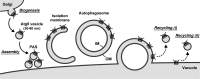
Comment in
-
Autophagy regulation through Atg9 traffic.J Cell Biol. 2012 Jul 23;198(2):151-3. doi: 10.1083/jcb.201206119. J Cell Biol. 2012. PMID: 22826119 Free PMC article.
References
-
- Anderson C.M., Georgiou G.N., Morrison I.E., Stevenson G.V., Cherry R.J. 1992. Tracking of cell surface receptors by fluorescence digital imaging microscopy using a charge-coupled device camera. Low-density lipoprotein and influenza virus receptor mobility at 4 degrees C. J. Cell Sci. 101:415–425 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

