Amino acid starvation induces reactivation of silenced transgenes and latent HIV-1 provirus via down-regulation of histone deacetylase 4 (HDAC4)
- PMID: 22826225
- PMCID: PMC3427085
- DOI: 10.1073/pnas.1202174109
Amino acid starvation induces reactivation of silenced transgenes and latent HIV-1 provirus via down-regulation of histone deacetylase 4 (HDAC4)
Abstract
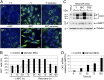
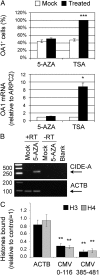
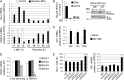
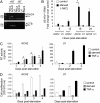
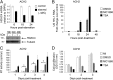
The epigenetic silencing of exogenous transcriptional units integrated into the genome represents a critical problem both for long-term gene therapy efficacy and for the eradication of latent viral infections. We report here that limitation of essential amino acids, such as methionine and cysteine, causes selective up-regulation of exogenous transgene expression in mammalian cells. Prolonged amino acid deprivation led to significant and reversible increase in the expression levels of stably integrated transgenes transcribed by means of viral or human promoters in HeLa cells. This phenomenon was mediated by epigenetic chromatin modifications, because histone deacetylase (HDAC) inhibitors reproduced starvation-induced transgene up-regulation, and transcriptome analysis, ChIP, and pharmacological and RNAi approaches revealed that a specific class II HDAC, namely HDAC4, plays a critical role in maintaining the silencing of exogenous transgenes. This mechanism was also operational in cells chronically infected with HIV-1, the etiological agent of AIDS, in a latency state. Indeed, both amino acid starvation and pharmacological inhibition of HDAC4 promoted reactivation of HIV-1 transcription and reverse transcriptase activity production in HDAC4(+) ACH-2 T-lymphocytic cells but not in HDAC4(-) U1 promonocytic cells. Thus, amino acid deprivation leads to transcriptional derepression of silenced transgenes, including integrated plasmids and retroviruses, by a process involving inactivation or down-regulation of HDAC4. These findings suggest that selective targeting of HDAC4 might represent a unique strategy for modulating the expression of therapeutic viral vectors, as well as that of integrated HIV-1 proviruses in latent reservoirs without significant cytotoxicity.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
Comment in
-
Viral infection: Coaxing HIV out of hiding.Nat Rev Microbiol. 2012 Sep;10(9):596-7. doi: 10.1038/nrmicro2861. Epub 2012 Aug 13. Nat Rev Microbiol. 2012. PMID: 22886238 No abstract available.
References
-
- Fischle W, et al. Enzymatic activity associated with class II HDACs is dependent on a multiprotein complex containing HDAC3 and SMRT/N-CoR. Mol Cell. 2002;9:45–57. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

