Optimization and comparison of knockdown efficacy between polymerase II expressed shRNA and artificial miRNA targeting luciferase and Apolipoprotein B100
- PMID: 22827812
- PMCID: PMC3424168
- DOI: 10.1186/1472-6750-12-42
Optimization and comparison of knockdown efficacy between polymerase II expressed shRNA and artificial miRNA targeting luciferase and Apolipoprotein B100
Abstract
Background: Controlling and limiting the expression of short hairpin RNA (shRNA) by using constitutive or tissue-specific polymerase II (pol II) expression can be a promising strategy to avoid RNAi toxicity. However, to date detailed studies on requirements for effective pol II shRNA expression and processing are not available. We investigated the optimal structural configuration of shRNA molecules, namely: hairpin location, stem length and termination signal required for effective pol II expression and compared it with an alternative strategy of avoiding toxicity by using artificial microRNA (miRNA) scaffolds.
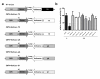
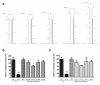
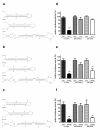
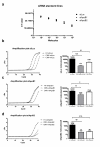
Results: Highly effective shRNAs targeting luciferase (shLuc) or Apolipoprotein B100 (shApoB1 and shApoB2) were placed under the control of the pol II CMV promoter and expressed at +5 or +6 nucleotides (nt) with reference to the transcription start site (TSS). Different transcription termination signals (TTS), namely minimal polyadenylation (pA), poly T (T5) and U1 were also used. All pol II- expressed shRNA variants induced mild inhibition of Luciferase reporters carrying specific targets and none of them showed comparable efficacy to their polymerase III-expressed H1-shRNA controls, regardless of hairpin position and termination signal used. Extending hairpin stem length from 20 basepairs (bp) to 21, 25 or 29 bp yielded only slight improvement in the overall efficacy. When shLuc, shApoB1 and shApoB2 were placed in an artificial miRNA scaffold, two out of three were as potent as the H1-shRNA controls. Quantification of small interfering RNA (siRNA) molecules showed that the artificial miRNA constructs expressed less molecules than H1-shRNAs and that CMV-shRNA expressed the lowest amount of siRNA molecules suggesting that RNAi processing in this case is least effective. Furthermore, CMV-miApoB1 and CMV-miApoB2 were as effective as the corresponding H1-shApoB1 and H1-shApoB2 in inhibiting endogenous ApoB mRNA.
Conclusion: Our results demonstrate that artificial miRNA have a better efficacy profile than shRNA expressed either from H1 or CMV promoter and will be used in the future for RNAi therapeutic development.
Figures
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

