Analysis of genomic aberrations and gene expression profiling identifies novel lesions and pathways in myeloproliferative neoplasms
- PMID: 22829077
- PMCID: PMC3256752
- DOI: 10.1038/bcj.2011.39
Analysis of genomic aberrations and gene expression profiling identifies novel lesions and pathways in myeloproliferative neoplasms
Abstract
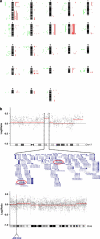
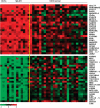
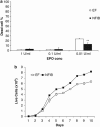
Polycythemia vera (PV), essential thrombocythemia and primary myelofibrosis, are myeloproliferative neoplasms (MPNs) with distinct clinical features and are associated with the JAK2V617F mutation. To identify genomic anomalies involved in the pathogenesis of these disorders, we profiled 87 MPN patients using Affymetrix 250K single-nucleotide polymorphism (SNP) arrays. Aberrations affecting chr9 were the most frequently observed and included 9pLOH (n=16), trisomy 9 (n=6) and amplifications of 9p13.3-23.3 (n=1), 9q33.1-34.13 (n=1) and 9q34.13 (n=6). Patients with trisomy 9 were associated with elevated JAK2V617F mutant allele burden, suggesting that gain of chr9 represents an alternative mechanism for increasing JAK2V617F dosage. Gene expression profiling of patients with and without chr9 abnormalities (+9, 9pLOH), identified genes potentially involved in disease pathogenesis including JAK2, STAT5B and MAPK14. We also observed recurrent gains of 1p36.31-36.33 (n=6), 17q21.2-q21.31 (n=5) and 17q25.1-25.3 (n=5) and deletions affecting 18p11.31-11.32 (n=8). Combined SNP and gene expression analysis identified aberrations affecting components of a non-canonical PRC2 complex (EZH1, SUZ12 and JARID2) and genes comprising a 'HSC signature' (MLLT3, SMARCA2 and PBX1). We show that NFIB, which is amplified in 7/87 MPN patients and upregulated in PV CD34+ cells, protects cells from apoptosis induced by cytokine withdrawal.
Figures

Similar articles
-
Genomic aberrations of myeloproliferative and myelodysplastic/myeloproliferative neoplasms in chronic phase and during disease progression.Int J Lab Hematol. 2015 Apr;37(2):181-9. doi: 10.1111/ijlh.12257. Epub 2014 May 21. Int J Lab Hematol. 2015. PMID: 24845343
-
Clonal analyses define the relationships between chromosomal abnormalities and JAK2V617F in patients with Ph-negative myeloproliferative neoplasms.Exp Hematol. 2009 Oct;37(10):1194-200. doi: 10.1016/j.exphem.2009.07.003. Epub 2009 Jul 15. Exp Hematol. 2009. PMID: 19615425
-
Proinflammatory Cytokine IL-6 and JAK-STAT Signaling Pathway in Myeloproliferative Neoplasms.Mediators Inflamm. 2015;2015:453020. doi: 10.1155/2015/453020. Epub 2015 Sep 29. Mediators Inflamm. 2015. PMID: 26491227 Free PMC article.
-
Clinical correlates of JAK2V617F presence or allele burden in myeloproliferative neoplasms: a critical reappraisal.Leukemia. 2008 Jul;22(7):1299-307. doi: 10.1038/leu.2008.113. Epub 2008 May 22. Leukemia. 2008. PMID: 18496562 Review.
-
JAK2 and MPL mutations in myeloproliferative neoplasms: discovery and science.Leukemia. 2008 Oct;22(10):1813-7. doi: 10.1038/leu.2008.229. Epub 2008 Aug 28. Leukemia. 2008. PMID: 18754026 Review.
Cited by
-
MAPK14 over-expression is a transcriptomic feature of polycythemia vera and correlates with adverse clinical outcomes.J Transl Med. 2021 May 31;19(1):233. doi: 10.1186/s12967-021-02913-3. J Transl Med. 2021. PMID: 34059095 Free PMC article.
-
The Positive Correlations between the Expression of Histopathological Ubiquitin-Conjugating Enzyme 2O Staining and Prostate Cancer Advancement.Pharmaceuticals (Basel). 2021 Aug 8;14(8):778. doi: 10.3390/ph14080778. Pharmaceuticals (Basel). 2021. PMID: 34451875 Free PMC article.
-
Patients with triple-negative, JAK2V617F- and CALR-mutated essential thrombocythemia share a unique gene expression signature.Blood Adv. 2021 Feb 23;5(4):1059-1068. doi: 10.1182/bloodadvances.2020003172. Blood Adv. 2021. PMID: 33599741 Free PMC article.
-
Establishment of the TALE-code reveals aberrantly activated homeobox gene PBX1 in Hodgkin lymphoma.PLoS One. 2021 Feb 4;16(2):e0246603. doi: 10.1371/journal.pone.0246603. eCollection 2021. PLoS One. 2021. PMID: 33539429 Free PMC article.
-
Reduced Expression of Jumonji AT-Rich Interactive Domain 2 (JARID2) in Glioma Inhibits Tumor Growth In Vitro and In Vivo.Oncol Res. 2017 Mar 13;25(3):365-372. doi: 10.3727/096504016X14738135889976. Epub 2016 Sep 16. Oncol Res. 2017. PMID: 27641964 Free PMC article.
References
-
- Delhommeau F, Jeziorowska D, Marzac C, Casadevall N. Molecular aspects of myeloproliferative neoplasms. Int J Hematol. 2010;91:165–173. - PubMed
-
- Ihle JN, Gilliland DG. Jak2: normal function and role in hematopoietic disorders. Curr Opin Genet Dev. 2007;17:8–14. - PubMed
-
- Liu KD, Gaffen SL, Goldsmith MA. JAK/STAT signaling by cytokine receptors. Curr Opin Immunol. 1998;10:271–278. - PubMed
-
- Morgan KJ, Gilliland DG. A role for JAK2 mutations in myeloproliferative diseases. Annu Rev Med. 2008;59:213–222. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

