Gene expression signatures associated with the in vitro resistance to two tyrosine kinase inhibitors, nilotinib and imatinib
- PMID: 22829191
- PMCID: PMC3255246
- DOI: 10.1038/bcj.2011.32
Gene expression signatures associated with the in vitro resistance to two tyrosine kinase inhibitors, nilotinib and imatinib
Abstract
The use of selective inhibitors targeting Bcr-Abl kinase is now established as a standard protocol in the treatment of chronic myelogenous leukemia; however, the acquisition of drug resistance is a major obstacle limiting the treatment efficacy. To elucidate the molecular mechanism of drug resistance, we established K562 cell line models resistant to nilotinib and imatinib. Microarray-based transcriptome profiling of resistant cells revealed that nilotinib- and imatinib-resistant cells showed the upregulation of kinase-encoding genes (AURKC, FYN, SYK, BTK and YES1). Among them, the upregulation of AURKC and FYN was observed both in nilotinib- and imatinib-resistant cells irrespective of exposure doses, while SYK, BTK and YES1 showed dose-dependent upregulation of expression. Upregulation of EGF and JAG1 oncogenes as well as genes encoding ATP-dependent drug efflux pump proteins such as ABCB1 was also observed in the resistant cells, which may confer alternative survival benefits. Functional gene set analysis revealed that molecular categories of 'ATPase activity', 'cell adhesion' or 'tyrosine kinase activity' were commonly activated in the resistant clones. Taken together, the transcriptome analysis of tyrosine kinase inhibitors (TKI)-resistant clones provides the insights into the mechanism of drug resistance, which can facilitate the development of an effective screening method as well as therapeutic intervention to deal with TKI resistance.
Figures



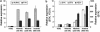
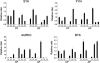
Similar articles
-
Expression of LYN and PTEN genes in chronic myeloid leukemia and their importance in therapeutic strategy.Blood Cells Mol Dis. 2014 Feb-Mar;52(2-3):121-5. doi: 10.1016/j.bcmd.2013.09.002. Epub 2013 Oct 3. Blood Cells Mol Dis. 2014. PMID: 24091144
-
A molecular and biophysical comparison of macromolecular changes in imatinib-sensitive and imatinib-resistant K562 cells exposed to ponatinib.Tumour Biol. 2016 Feb;37(2):2365-78. doi: 10.1007/s13277-015-4015-9. Epub 2015 Sep 15. Tumour Biol. 2016. PMID: 26373734
-
Gene expression profiling of imatinib and PD166326-resistant CML cell lines identifies Fyn as a gene associated with resistance to BCR-ABL inhibitors.Mol Cancer Ther. 2009 Jul;8(7):1924-33. doi: 10.1158/1535-7163.MCT-09-0168. Epub 2009 Jun 30. Mol Cancer Ther. 2009. PMID: 19567819
-
Important therapeutic targets in chronic myelogenous leukemia.Clin Cancer Res. 2007 Feb 15;13(4):1089-97. doi: 10.1158/1078-0432.CCR-06-2147. Clin Cancer Res. 2007. PMID: 17317816 Review.
-
Optimizing outcomes for patients with advanced disease in chronic myelogenous leukemia.Semin Oncol. 2008 Feb;35(1 Suppl 1):S1-17; quiz S18-20. doi: 10.1053/j.seminoncol.2007.12.002. Semin Oncol. 2008. PMID: 18346528 Review.
Cited by
-
Systematic prediction of drug resistance caused by transporter genes in cancer cells.Sci Rep. 2021 Apr 1;11(1):7400. doi: 10.1038/s41598-021-86921-9. Sci Rep. 2021. PMID: 33795761 Free PMC article.
-
Profiling the Expression and Prognostic Values of FYN, A Non-Receptor Tyrosine Kinase, in Different Histological Types of Epithelial Ovarian Cancer.Asian Pac J Cancer Prev. 2023 Jan 1;24(1):321-329. doi: 10.31557/APJCP.2023.24.1.321. Asian Pac J Cancer Prev. 2023. PMID: 36708583 Free PMC article.
-
Role of YES1 signaling in tumor therapy resistance.Cancer Innov. 2023 Mar 3;2(3):210-218. doi: 10.1002/cai2.51. eCollection 2023 Jun. Cancer Innov. 2023. PMID: 38089407 Free PMC article. Review.
-
lncRNA and Mechanisms of Drug Resistance in Cancers of the Genitourinary System.Cancers (Basel). 2020 Aug 3;12(8):2148. doi: 10.3390/cancers12082148. Cancers (Basel). 2020. PMID: 32756406 Free PMC article. Review.
-
Network pharmacology strategies toward multi-target anticancer therapies: from computational models to experimental design principles.Curr Pharm Des. 2014;20(1):23-36. doi: 10.2174/13816128113199990470. Curr Pharm Des. 2014. PMID: 23530504 Free PMC article. Review.
References
-
- Hehlmann R, Hochhaus A, Baccarani M. Chronic myeloid leukaemia. Lancet. 2007;370:342–350. - PubMed
-
- Bartram CR, de Klein A, Hagemeijer A, van Agthoven T, Geurts van Kessel A, Bootsma D, et al. Translocation of c-ab1 oncogene correlates with the presence of a Philadelphia chromosome in chronic myelocytic leukaemia. Nature. 1983;306:277–280. - PubMed
-
- Deininger M, Buchdunger E, Druker BJ. The development of imatinib as a therapeutic agent for chronic myeloid leukemia. Blood. 2005;105:2640–2653. - PubMed
-
- Hochhaus A, O'Brien SG, Guilhot F, Druker BJ, Branford S, Foroni L, et al. Six-year follow-up of patients receiving imatinib for the first-line treatment of chronic myeloid leukemia. Leukemia. 2009;23:1054–1061. - PubMed
-
- O'Brien SG, Guilhot F, Larson RA, Gathmann I, Baccarani M, Cervantes F, et al. Imatinib compared with interferon and low-dose cytarabine for newly diagnosed chronic-phase chronic myeloid leukemia. N Engl J Med. 2003;348:994–1004. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

