Evidence of inbreeding depression on human height
- PMID: 22829771
- PMCID: PMC3400549
- DOI: 10.1371/journal.pgen.1002655
Evidence of inbreeding depression on human height
Abstract
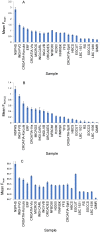
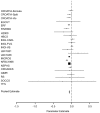
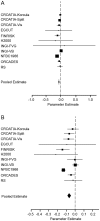
Stature is a classical and highly heritable complex trait, with 80%-90% of variation explained by genetic factors. In recent years, genome-wide association studies (GWAS) have successfully identified many common additive variants influencing human height; however, little attention has been given to the potential role of recessive genetic effects. Here, we investigated genome-wide recessive effects by an analysis of inbreeding depression on adult height in over 35,000 people from 21 different population samples. We found a highly significant inverse association between height and genome-wide homozygosity, equivalent to a height reduction of up to 3 cm in the offspring of first cousins compared with the offspring of unrelated individuals, an effect which remained after controlling for the effects of socio-economic status, an important confounder (χ(2) = 83.89, df = 1; p = 5.2 × 10(-20)). There was, however, a high degree of heterogeneity among populations: whereas the direction of the effect was consistent across most population samples, the effect size differed significantly among populations. It is likely that this reflects true biological heterogeneity: whether or not an effect can be observed will depend on both the variance in homozygosity in the population and the chance inheritance of individual recessive genotypes. These results predict that multiple, rare, recessive variants influence human height. Although this exploratory work focuses on height alone, the methodology developed is generally applicable to heritable quantitative traits (QT), paving the way for an investigation into inbreeding effects, and therefore genetic architecture, on a range of QT of biomedical importance.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
References
-
- Cole TJ. The secular trend in human physical growth: a biological view. Econ Hum Biol. 2003;1:161–168. - PubMed
-
- Ogden CL, Fryar CD, Carroll MD, Flegal KM. Mean body weight, height, and body mass index, United States 1960–2002. Adv Data. 2004:1–17. - PubMed
-
- Macgregor S, Cornes BK, Martin NG, Visscher PM. Bias, precision and heritability of self-reported and clinically measured height in Australian twins. Hum Genet. 2006;120:571–580. - PubMed
-
- Preece MA. The genetic contribution to stature. Horm Res. 1996;45:56–58. - PubMed
Publication types
MeSH terms
Grants and funding
- K05 AA017688/AA/NIAAA NIH HHS/United States
- MC_PC_U127527180/MRC_/Medical Research Council/United Kingdom
- MC_U127527198/MRC_/Medical Research Council/United Kingdom
- G0701120/MRC_/Medical Research Council/United Kingdom
- G0700704/MRC_/Medical Research Council/United Kingdom
- BB/F019394/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- ETM/55/CSO_/Chief Scientist Office/United Kingdom
- CZB/4/505/CSO_/Chief Scientist Office/United Kingdom
- CZB/4/710/CSO_/Chief Scientist Office/United Kingdom
- MC_U127561128/MRC_/Medical Research Council/United Kingdom
- 12076/CRUK_/Cancer Research UK/United Kingdom
- 090532/WT_/Wellcome Trust/United Kingdom
- MC_PC_U127561128/MRC_/Medical Research Council/United Kingdom
LinkOut - more resources
Full Text Sources
Miscellaneous

