The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism
- PMID: 22829779
- PMCID: PMC3400554
- DOI: 10.1371/journal.pgen.1002816
The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism
Abstract
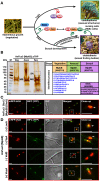
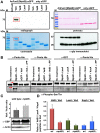
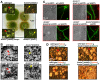
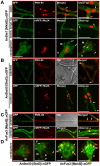
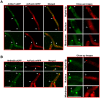
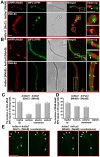
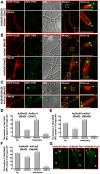
The sexual Fus3 MAP kinase module of yeast is highly conserved in eukaryotes and transmits external signals from the plasma membrane to the nucleus. We show here that the module of the filamentous fungus Aspergillus nidulans (An) consists of the AnFus3 MAP kinase, the upstream kinases AnSte7 and AnSte11, and the AnSte50 adaptor. The fungal MAPK module controls the coordination of fungal development and secondary metabolite production. It lacks the membrane docking yeast Ste5 scaffold homolog; but, similar to yeast, the entire MAPK module's proteins interact with each other at the plasma membrane. AnFus3 is the only subunit with the potential to enter the nucleus from the nuclear envelope. AnFus3 interacts with the conserved nuclear transcription factor AnSte12 to initiate sexual development and phosphorylates VeA, which is a major regulatory protein required for sexual development and coordinated secondary metabolite production. Our data suggest that not only Fus3, but even the entire MAPK module complex of four physically interacting proteins, can migrate from plasma membrane to nuclear envelope.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
-
- Marshall CJ. MAP kinase kinase kinase, MAP kinase kinase and MAP kinase. Curr Opin Genet Dev. 1994;4:82–89. - PubMed
-
- Roman E, Arana DM, Nombela C, Alonso-Monge R, Pla J. MAP kinase pathways as regulators of fungal virulence. Trends Microbiol. 2007;15:181–190. - PubMed
-
- Saito H. Regulation of cross-talk in yeast MAPK signaling pathways. Curr Opin Microbiol. 2010;13:677–683. - PubMed
-
- Bardwell L. A walk-through of the yeast mating pheromone response pathway. Peptides. 2004;25:1465–1476. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

