Cre-mediated stress affects sirtuin expression levels, peroxisome biogenesis and metabolism, antioxidant and proinflammatory signaling pathways
- PMID: 22829911
- PMCID: PMC3400606
- DOI: 10.1371/journal.pone.0041097
Cre-mediated stress affects sirtuin expression levels, peroxisome biogenesis and metabolism, antioxidant and proinflammatory signaling pathways
Abstract
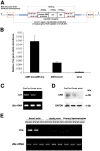
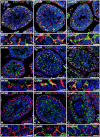
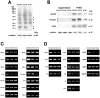
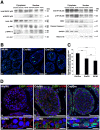
Cre-mediated excision of loxP sites is widely used in mice to manipulate gene function in a tissue-specific manner. To analyze phenotypic alterations related to Cre-expression, we have used AMH-Cre-transgenic mice as a model system. Different Cre expression levels were obtained by investigation of C57BL/6J wild type as well as heterozygous and homozygous AMH-Cre-mice. Our results indicate that Cre-expression itself in Sertoli cells already has led to oxidative stress and lipid peroxidation (4-HNE lysine adducts), inducing PPARα/γ, peroxisome proliferation and alterations of peroxisome biogenesis (PEX5, PEX13 and PEX14) as well as metabolic proteins (ABCD1, ABCD3, MFP1, thiolase B, catalase). In addition to the strong catalase increase, a NRF2- and FOXO3-mediated antioxidative response (HMOX1 of the endoplasmic reticulum and mitochondrial SOD2) and a NF-κB activation were noted. TGFβ1 and proinflammatory cytokines like IL1, IL6 and TNFα were upregulated and stress-related signaling pathways were induced. Sertoli cell mRNA-microarray analysis revealed an increase of TNFR2-signaling components. 53BP1 recruitment and expression levels for DNA repair genes as well as for p53 were elevated and the ones for related sirtuin deacetylases affected (SIRT 1, 3-7) in Sertoli cells. Under chronic Cre-mediated DNA damage conditions a strong downregulation of Sirt1 was observed, suggesting that the decrease of this important coordinator between DNA repair and metabolic signaling might induce the repression release of major transcription factors regulating metabolic and cytokine-mediated stress pathways. Indeed, caspase-3 was activated and increased germ cell apoptosis was observed, suggesting paracrine effects. In conclusion, the observed wide stress-induced effects and metabolic alterations suggest that it is essential to use the correct control animals (Cre/Wt) with matched Cre expression levels to differentiate between Cre-mediated and specific gene-knock out-mediated effects.
Conflict of interest statement
Figures



References
-
- Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. - PubMed
-
- Lobe CG, Nagy A. Conditional genome alteration in mice. Bioessays. 1998;20:200–208. - PubMed
-
- Sternberg N, Hamilton D. Bacteriophage P1 site-specific recombination. I. Recombination between loxP sites. J Mol Biol. 1981;150:467–486. - PubMed
-
- Lewandoski M. Conditional control of gene expression in the mouse. Nat Rev Genet. 2001;2:743–755. - PubMed
-
- Furuta Y, Behringer RR. Recent innovations in tissue-specific gene modifications in the mouse. Birth Defects Res C Embryo Today. 2005;75:43–57. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

