Luminal A and luminal B (HER2 negative) subtypes of breast cancer consist of a mixture of tumors with different genotype
- PMID: 22830453
- PMCID: PMC3413599
- DOI: 10.1186/1756-0500-5-376
Luminal A and luminal B (HER2 negative) subtypes of breast cancer consist of a mixture of tumors with different genotype
Abstract
Background: The St Gallen International Expert Consensus 2011 has proposed a new classification system for breast cancer. The purpose of this study was to elucidate the relationship between the breast cancer subtypes determined by the new classification system and genomic characteristics.
Methods: Invasive breast cancers (n = 363) were immunohistochemically classified as follows: 111 (30.6%) as luminal A, 95 (26.2%) as luminal B (HER2 negative), 69 (19.0%) as luminal B (HER2 positive), 41 (11.3%) as HER2, and 47 (12.9%) as basal-like subtypes.
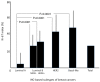
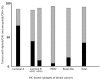
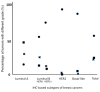
Results: The high expression of Ki-67 antigen was detected in 236 tumors; no cases of luminal A subtype showed high expression of the Ki-67 antigen, but more than 85% of tumors of the other subtypes showed high expression. In addition, DNA ploidy and chromosomal instability (CIN) were assessed using imaging cytometry and FISH, respectively. In this series, 336 (92.6%) tumors consisted of 129 diploid/CIN- and 207 aneuploid/CIN + tumors. Diploid/CIN- and aneuploid/CIN+ features were detected in 64.9% and 27.9% of luminal A, 41.1% and 49.5% of luminal B (HER2-), 11.6% and 81.2% of luminal B (HER2+), 4.9% and 90.2% of HER2, and 17.0% and 76.6% of basal-like subtypes, respectively. Unlike the luminal B (HER2+), HER2 and basal-like subtypes, the luminal A and luminal B (HER2-) subtypes were heterogeneous in terms of DNA ploidy and CIN.
Conclusions: It is reasonable to propose that the luminal A and luminal B (HER2-) subtypes should be further divided into two subgroups, diploid/CIN- and aneuploid/CIN+, based on their underlying genomic status.
Figures


References
-
- Raica M, Jung I, Cimpean AM, Suciu C, Muresan AM. From conventional pathologic diagnosis to the molecular classification of breast carcinoma: are we ready for the change? Rpm J Mrphol Embryol. 2009;50:5–13. - PubMed
-
- Goldhirsch A, Wood WC, Coates AS, Gelber RD, Thürlimann B, Senn HJ. Panel members. Strategies for subtypes--dealing with the diversity of breast cancer: highlights of the St. Gallen international expert consensus on the primary therapy of early breast cancer 2011. Ann Oncol. 2011;22:1736–1747. doi: 10.1093/annonc/mdr304. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

