Developmental features of DNA methylation during activation of the embryonic zebrafish genome
- PMID: 22830626
- PMCID: PMC3491385
- DOI: 10.1186/gb-2012-13-7-r65
Developmental features of DNA methylation during activation of the embryonic zebrafish genome
Abstract
Background: Zygotic genome activation (ZGA) occurs at the mid-blastula transition (MBT) in zebrafish and is a period of extensive chromatin remodeling. Genome-scale gametic demethylation and remethylation occurs after fertilization, during blastula stages, but how ZGA relates to promoter DNA methylation states is unknown. Using methylated DNA immunoprecipitation coupled to high-density microarray hybridization, we characterize genome-wide promoter DNA methylation dynamics before, during and after ZGA onset, in relation to changes in post-translational histone modifications and gene expression.
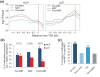
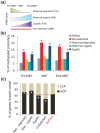
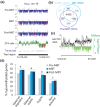
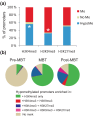
Results: We show methylation of thousands of promoters before ZGA and additional methylation after ZGA, finding more dynamic methylation -1 to 0 kb upstream of the transcription start site than downstream. The MBT is marked by differential methylation of high and low CpG promoters, and we identify hypomethylated promoters that are mostly CG-rich and remain hypomethylated through the MBT. Hypomethylated regions constitute a platform for H3K4me3, whereas H3K9me3 preferentially associates with methylated regions. H3K27me3 associates with either methylation state depending on its coincidence with H3K4me3 or H3K9me3. Cohorts of genes differentially expressed through the MBT period display distinct promoter methylation patterns related to CG content rather than transcriptional fate. Lastly, although a significant proportion of genes methylated in sperm are unmethylated in embryos, over 90% of genes methylated in embryos are also methylated in sperm.
Conclusions: Our results suggest a pre-patterning of developmental gene expression potential by a combination of DNA hypomethylation and H3K4 trimethylation on CG-rich promoters, and are consistent with a transmission of DNA methylation states from gametes to early embryos.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

