Development and validation of a high-content screening assay to identify inhibitors of cytoplasmic dynein-mediated transport of glucocorticoid receptor to the nucleus
- PMID: 22830992
- PMCID: PMC3464420
- DOI: 10.1089/adt.2012.456
Development and validation of a high-content screening assay to identify inhibitors of cytoplasmic dynein-mediated transport of glucocorticoid receptor to the nucleus
Abstract
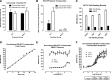
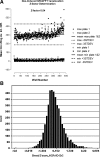
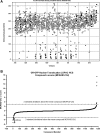
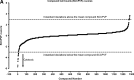
Rapid ligand-induced trafficking of glucocorticoid nuclear hormone receptor (GR) from the cytoplasm to the nucleus is an extensively studied model for intracellular retrograde cargo transport employed in constructive morphogenesis and many other cellular functions. Unfortunately, potent and selective small-molecule disruptors of this process are lacking, which has restricted pharmacological investigations. We describe here the development and validation of a 384-well high-content screening (HCS) assay to identify inhibitors of the rapid ligand-induced retrograde translocation of cytoplasmic glucocorticoid nuclear hormone receptor green fluorescent fusion protein (GR-GFP) into the nuclei of 3617.4 mouse mammary adenocarcinoma cells. We selected 3617.4 cells, because they express GR-GFP under the control of a tetracycline (Tet)-repressible promoter and are exceptionally amenable to image acquisition and analysis procedures. Initially, we investigated the time-dependent expression of GR-GFP in 3617.4 cells under Tet-on and Tet-off control to determine the optimal conditions to measure dexamethasone (Dex)-induced GR-GFP nuclear translocation on the ArrayScan-VTI automated imaging platform. We then miniaturized the assay into a 384-well format and validated the performance of the GR-GFP nuclear translocation HCS assay in our 3-day assay signal window and dimethylsulfoxide validation tests. The molecular chaperone heat shock protein 90 (Hsp90) plays an essential role in the regulation of GR steroid binding affinity and ligand-induced retrograde trafficking to the nucleus. We verified that the GR-GFP HCS assay captured the concentration-dependent inhibition of GR-GFP nuclear translocation by 17-AAG, a benzoquinone ansamycin that selectively blocks the binding and hydrolysis of ATP by Hsp90. We screened the 1280 compound library of pharmacologically active compounds set in the Dex-induced GR-GFP nuclear translocation assay and used the multi-parameter HCS data to eliminate cytotoxic compounds and fluorescent outliers. We identified five qualified hits that inhibited the rapid retrograde trafficking of GR-GFP in a concentration-dependent manner: Bay 11-7085, 4-phenyl-3-furoxancarbonitrile, parthenolide, apomorphine, and 6-nitroso-1,2-benzopyrone. The data presented here demonstrate that the GR-GFP HCS assay provides an effective phenotypic screen and support the proposition that screening a larger library of diversity compounds will yield novel small-molecule probes that will enable the further exploration of intracellular retrograde transport of cargo along microtubules, a process which is essential to the morphogenesis and function of all cells.
Figures













References
-
- Hirokawa N. Kinesin and dynein superfamily proteins and the mechanism of organelle transport. Science. 1998;279:519–526. - PubMed
-
- Hirokawa N. Noda Y. Tanaka Y. Niwa S. Kinesin superfamily motor proteins and intracellular transport. Nat Rev Mol Cell Biol. 2009;10:682–696. - PubMed
-
- Vallee R. Williams JC. Varma D. Barnhart LE. Dynein: an ancient motor protein involved in multiple modes of transport. J Neurobiol. 2003;58:189–200. - PubMed
-
- Vallee R. Höök P. Autoinhibitory and other autoregulatory elements within the dynein motor domain. J Struct Biol. 2006;156:175–181. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

