Premetazoan origin of the hippo signaling pathway
- PMID: 22832104
- PMCID: PMC3406323
- DOI: 10.1016/j.celrep.2011.11.004
Premetazoan origin of the hippo signaling pathway
Abstract
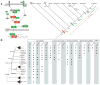
Nonaggregative multicellularity requires strict control of cell number. The Hippo signaling pathway coordinates cell proliferation and apoptosis and is a central regulator of organ size in animals. Recent studies have shown the presence of key members of the Hippo pathway in nonbilaterian animals, but failed to identify this pathway outside Metazoa. Through comparative analyses of recently sequenced holozoan genomes, we show that Hippo pathway components, such as the kinases Hippo and Warts, the coactivator Yorkie, and the transcription factor Scalloped, were already present in the unicellular ancestors of animals. Remarkably, functional analysis of Hippo components of the amoeboid holozoan Capsaspora owczarzaki, performed in Drosophila melanogaster, demonstrate that the growth-regulatory activity of the Hippo pathway is conserved in this unicellular lineage. Our findings show that the Hippo pathway evolved well before the origin of Metazoa and highlight the importance of Hippo signaling as a key developmental mechanism predating the origin of Metazoa.
Copyright © 2012 The Authors. Published by Elsevier Inc. All rights reserved.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

