Reverse engineering biomolecular systems using -omic data: challenges, progress and opportunities
- PMID: 22833495
- PMCID: PMC3404400
- DOI: 10.1093/bib/bbs026
Reverse engineering biomolecular systems using -omic data: challenges, progress and opportunities
Abstract
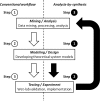
Recent advances in high-throughput biotechnologies have led to the rapid growing research interest in reverse engineering of biomolecular systems (REBMS). 'Data-driven' approaches, i.e. data mining, can be used to extract patterns from large volumes of biochemical data at molecular-level resolution while 'design-driven' approaches, i.e. systems modeling, can be used to simulate emergent system properties. Consequently, both data- and design-driven approaches applied to -omic data may lead to novel insights in reverse engineering biological systems that could not be expected before using low-throughput platforms. However, there exist several challenges in this fast growing field of reverse engineering biomolecular systems: (i) to integrate heterogeneous biochemical data for data mining, (ii) to combine top-down and bottom-up approaches for systems modeling and (iii) to validate system models experimentally. In addition to reviewing progress made by the community and opportunities encountered in addressing these challenges, we explore the emerging field of synthetic biology, which is an exciting approach to validate and analyze theoretical system models directly through experimental synthesis, i.e. analysis-by-synthesis. The ultimate goal is to address the present and future challenges in reverse engineering biomolecular systems (REBMS) using integrated workflow of data mining, systems modeling and synthetic biology.
Figures

Similar articles
-
Engineering with Biomolecular Motors.Acc Chem Res. 2018 Dec 18;51(12):3015-3022. doi: 10.1021/acs.accounts.8b00296. Epub 2018 Oct 30. Acc Chem Res. 2018. PMID: 30376292
-
Are the biomedical sciences ready for synthetic biology?Biomol Concepts. 2020 Jan 24;11(1):23-31. doi: 10.1515/bmc-2020-0003. Biomol Concepts. 2020. PMID: 31982863 Review.
-
Modular design: Implementing proven engineering principles in biotechnology.Biotechnol Adv. 2019 Nov 15;37(7):107403. doi: 10.1016/j.biotechadv.2019.06.002. Epub 2019 Jun 7. Biotechnol Adv. 2019. PMID: 31181317 Review.
-
An engineering design approach to systems biology.Integr Biol (Camb). 2017 Jul 17;9(7):574-583. doi: 10.1039/c7ib00014f. Integr Biol (Camb). 2017. PMID: 28590470 Free PMC article. Review.
-
Can biological complexity be reverse engineered?Stud Hist Philos Biol Biomed Sci. 2015 Oct;53:73-83. doi: 10.1016/j.shpsc.2015.03.008. Epub 2015 Apr 19. Stud Hist Philos Biol Biomed Sci. 2015. PMID: 25903121
Cited by
-
A four-lncRNA risk signature for prognostic prediction of osteosarcoma.Front Genet. 2023 Jan 4;13:1081478. doi: 10.3389/fgene.2022.1081478. eCollection 2022. Front Genet. 2023. PMID: 36685868 Free PMC article.
-
Genetic association of ANRIL with susceptibility to Ischemic stroke: A comprehensive meta-analysis.PLoS One. 2022 Jun 2;17(6):e0263459. doi: 10.1371/journal.pone.0263459. eCollection 2022. PLoS One. 2022. PMID: 35653368 Free PMC article.
-
Genome sequencing of high-penicillin producing industrial strain of Penicillium chrysogenum.BMC Genomics. 2014;15 Suppl 1(Suppl 1):S11. doi: 10.1186/1471-2164-15-S1-S11. Epub 2014 Jan 24. BMC Genomics. 2014. PMID: 24564352 Free PMC article.
-
PLAU inferred from a correlation network is critical for suppressor function of regulatory T cells.Mol Syst Biol. 2012;8:624. doi: 10.1038/msb.2012.56. Mol Syst Biol. 2012. PMID: 23169000 Free PMC article.
-
Factors associated with health intentions and behaviour among health checkup participants in Japan.Sci Rep. 2021 Oct 5;11(1):19761. doi: 10.1038/s41598-021-99303-y. Sci Rep. 2021. PMID: 34611263 Free PMC article.

