Regulatory consequences of gene translocation in bacteria
- PMID: 22833608
- PMCID: PMC3467084
- DOI: 10.1093/nar/gks694
Regulatory consequences of gene translocation in bacteria
Abstract
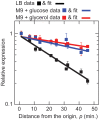
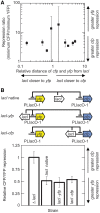
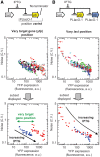
Gene translocations play an important role in the plasticity and evolution of bacterial genomes. In this study, we investigated the impact on gene regulation of three genome organizational features that can be altered by translocations: (i) chromosome position; (ii) gene orientation; and (iii) the distance between a target gene and its transcription factor gene ('target-TF distance'). Specifically, we quantified the effect of these features on constitutive expression, transcription factor binding and/or gene expression noise using a synthetic network in Escherichia coli composed of a transcription factor (LacI repressor) and its target gene (yfp). Here we show that gene regulation is generally robust to changes in chromosome position, gene orientation and target-TF distance. The only demonstrable effect was that chromosome position alters constitutive expression, due to changes in gene copy number and local sequence effects, and that this determines maximum and minimum expression levels. The results were incorporated into a mathematical model which was used to quantitatively predict the responses of a simple gene network to gene translocations; the predictions were confirmed experimentally. In summary, gene translocation can modulate constitutive gene expression levels due to changes in chromosome position but it has minimal impact on other facets of gene regulation.
Figures




Similar articles
-
Tracking Low-Copy Transcription Factors in Living Bacteria: The Case of the lac Repressor.Biophys J. 2017 Apr 11;112(7):1316-1327. doi: 10.1016/j.bpj.2017.02.028. Biophys J. 2017. PMID: 28402875 Free PMC article.
-
Gene location and DNA density determine transcription factor distributions in Escherichia coli.Mol Syst Biol. 2012;8:610. doi: 10.1038/msb.2012.42. Mol Syst Biol. 2012. PMID: 22968444 Free PMC article.
-
Minimal effect of gene clustering on expression in Escherichia coli.Genetics. 2013 Feb;193(2):453-65. doi: 10.1534/genetics.112.147199. Epub 2012 Dec 5. Genetics. 2013. PMID: 23222655 Free PMC article.
-
New insights into the regulatory networks of paralogous genes in bacteria.Microbiology (Reading). 2010 Jan;156(Pt 1):14-22. doi: 10.1099/mic.0.033266-0. Epub 2009 Oct 22. Microbiology (Reading). 2010. PMID: 19850620 Review.
-
Single molecule approaches to transcription factor kinetics in living cells.FEBS Lett. 2009 Dec 17;583(24):3979-83. doi: 10.1016/j.febslet.2009.11.035. FEBS Lett. 2009. PMID: 19925790 Review.
Cited by
-
A standardized genome architecture for bacterial synthetic biology (SEGA).Nat Commun. 2021 Oct 7;12(1):5876. doi: 10.1038/s41467-021-26155-5. Nat Commun. 2021. PMID: 34620865 Free PMC article.
-
Chromosomal Arrangement of Phosphorelay Genes Couples Sporulation and DNA Replication.Cell. 2015 Jul 16;162(2):328-337. doi: 10.1016/j.cell.2015.06.012. Epub 2015 Jul 9. Cell. 2015. PMID: 26165942 Free PMC article.
-
Intelligent host engineering for metabolic flux optimisation in biotechnology.Biochem J. 2021 Oct 29;478(20):3685-3721. doi: 10.1042/BCJ20210535. Biochem J. 2021. PMID: 34673920 Free PMC article. Review.
-
Engineered promoters enable constant gene expression at any copy number in bacteria.Nat Biotechnol. 2018 Apr;36(4):352-358. doi: 10.1038/nbt.4111. Epub 2018 Mar 19. Nat Biotechnol. 2018. PMID: 29553576
-
Targeted and Repetitive Chromosomal Integration Enables High-Level Heterologous Gene Expression in Lactobacillus casei.Appl Environ Microbiol. 2019 Apr 18;85(9):e00033-19. doi: 10.1128/AEM.00033-19. Print 2019 May 1. Appl Environ Microbiol. 2019. PMID: 30824448 Free PMC article.
References
-
- Chandler MG, Pritchard RH. The effect of gene concentration and relative gene dosage on gene output in Escherichia coli. Mol. Gen. Genet. 1975;138:127–141. - PubMed
-
- Sousa C, de Lorenzo V, Cebolla A. Modulation of gene expression through chromosomal positioning in Escherichia coli. Microbiology. 1997;143(Pt 6):2071–2078. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

