Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti
- PMID: 22833609
- PMCID: PMC3467087
- DOI: 10.1093/nar/gks700
Sequencing of the smallest Apicomplexan genome from the human pathogen Babesia microti
Abstract
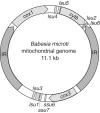
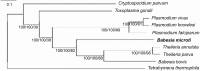
We have sequenced the genome of the emerging human pathogen Babesia microti and compared it with that of other protozoa. B. microti has the smallest nuclear genome among all Apicomplexan parasites sequenced to date with three chromosomes encoding ∼3500 polypeptides, several of which are species specific. Genome-wide phylogenetic analyses indicate that B. microti is significantly distant from all species of Babesidae and Theileridae and defines a new clade in the phylum Apicomplexa. Furthermore, unlike all other Apicomplexa, its mitochondrial genome is circular. Genome-scale reconstruction of functional networks revealed that B. microti has the minimal metabolic requirement for intraerythrocytic protozoan parasitism. B. microti multigene families differ from those of other protozoa in both the copy number and organization. Two lateral transfer events with significant metabolic implications occurred during the evolution of this parasite. The genomic sequencing of B. microti identified several targets suitable for the development of diagnostic assays and novel therapies for human babesiosis.
Figures


Similar articles
-
Sequence and annotation of the apicoplast genome of the human pathogen Babesia microti.PLoS One. 2014 Oct 3;9(10):e107939. doi: 10.1371/journal.pone.0107939. eCollection 2014. PLoS One. 2014. PMID: 25280009 Free PMC article.
-
Novel type of linear mitochondrial genomes with dual flip-flop inversion system in apicomplexan parasites, Babesia microti and Babesia rodhaini.BMC Genomics. 2012 Nov 14;13:622. doi: 10.1186/1471-2164-13-622. BMC Genomics. 2012. PMID: 23151128 Free PMC article.
-
Whole genome mapping and re-organization of the nuclear and mitochondrial genomes of Babesia microti isolates.PLoS One. 2013 Sep 4;8(9):e72657. doi: 10.1371/journal.pone.0072657. eCollection 2013. PLoS One. 2013. PMID: 24023759 Free PMC article.
-
Comparative Bioinformatics Analysis of Transcription Factor Genes Indicates Conservation of Key Regulatory Domains among Babesia bovis, Babesia microti, and Theileria equi.PLoS Negl Trop Dis. 2016 Nov 10;10(11):e0004983. doi: 10.1371/journal.pntd.0004983. eCollection 2016 Nov. PLoS Negl Trop Dis. 2016. PMID: 27832060 Free PMC article. Review.
-
Babesia sp.: emerging intracellular parasites in Europe.Pol J Microbiol. 2004;53 Suppl:55-60. Pol J Microbiol. 2004. PMID: 15787198 Review.
Cited by
-
Genome mining offers a new starting point for parasitology research.Parasitol Res. 2015 Feb;114(2):399-409. doi: 10.1007/s00436-014-4299-5. Epub 2015 Jan 8. Parasitol Res. 2015. PMID: 25563615 Review.
-
Genomic resources for a unique, low-virulence Babesia taxon from China.Parasit Vectors. 2016 Oct 27;9(1):564. doi: 10.1186/s13071-016-1846-1. Parasit Vectors. 2016. PMID: 27784333 Free PMC article.
-
Comparative and functional genomics of the protozoan parasite Babesia divergens highlighting the invasion and egress processes.PLoS Negl Trop Dis. 2019 Aug 19;13(8):e0007680. doi: 10.1371/journal.pntd.0007680. eCollection 2019 Aug. PLoS Negl Trop Dis. 2019. PMID: 31425518 Free PMC article.
-
Restriction enzyme digestion of host DNA enhances universal detection of parasitic pathogens in blood via targeted amplicon deep sequencing.Microbiome. 2018 Sep 17;6(1):164. doi: 10.1186/s40168-018-0540-2. Microbiome. 2018. PMID: 30223888 Free PMC article.
-
Severe babesiosis in immunocompetent man, Spain, 2011.Emerg Infect Dis. 2014 Apr;20(4):724-6. doi: 10.3201/eid2004.131409. Emerg Infect Dis. 2014. PMID: 24656155 Free PMC article. No abstract available.
References
-
- Hatcher JC, Greenberg PD, Antique J, Jimenez-Lucho VE. Severe babesiosis in Long Island: review of 34 cases and their complications. Clin. Infect. Dis. 2001;32:1117–1125. - PubMed
-
- Krause PJ, Gewurz BE, Hill D, Marty FM, Vannier E, Foppa IM, Furman RR, Neuhaus E, Skowron G, Gupta S, et al. Persistent and relapsing babesiosis in immunocompromised patients. Clin. Infect. Dis. 2008;46:370–376. - PubMed
-
- Institut of Medecine (US) Committee on Lyme Disease and Other Tick-borne Diseases: The State of the Science. Critical Needs and Gaps in Understanding Prevention, Amelioration, and Resolution of Lyme and Other Tick-Borne Diseases: The Short-Term and Long-Term Outcomes: Workshop Report. Washington, DC: National Academies Press; 2011. Prevention. In: Institute of Medicine of the National Academy of Sciences; pp. 155–176. - PubMed
-
- Hemmer RM, Ferrick DA, Conrad PA. Role of T cells and cytokines in fatal and resolving experimental babesiosis: protection in TNFRp55-/- mice infected with the human Babesia WA1 parasite. J. Parasitol. 2000;86:736–742. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

