Go hybrid: EM, crystallography, and beyond
- PMID: 22835744
- PMCID: PMC3478466
- DOI: 10.1016/j.sbi.2012.07.006
Go hybrid: EM, crystallography, and beyond
Abstract
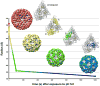
A mechanistic understanding of the molecular transactions that govern cellular function requires knowledge of the dynamic organization of the macromolecular machines involved in these processes. Structural biologists employ a variety of biophysical methods to study large macromolecular complexes, but no single technique is likely to provide a complete description of the structure-function relationship of all the constituent components. Since structural studies generally only provide snapshots of these dynamic machines as they accomplish their molecular functions, combining data from many methodologies is crucial to our understanding of molecular function.
Copyright © 2012 Elsevier Ltd. All rights reserved.
Figures

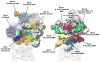

References
-
- Yonekura K, Maki-Yonekura S, Namba K. Complete atomic model of the bacterial flagellar filament by electron cryomicroscopy. Nature. 2003;424:643–650. - PubMed
-
- Becker T, Armache JP, Jarasch A, Anger AM, Villa E, Sieber H, Motaal BA, Mielke T, Berninghausen O, Beckmann R. Structure of the no-go mRNA decay complex Dom34-Hbs1 bound to a stalled 80S ribosome. Nat Struct Mol Biol. 2011;18:715–720. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

