Multiple binding of repressed mRNAs by the P-body protein Rck/p54
- PMID: 22836354
- PMCID: PMC3425784
- DOI: 10.1261/rna.034314.112
Multiple binding of repressed mRNAs by the P-body protein Rck/p54
Abstract
Translational repression is achieved by protein complexes that typically bind 3' UTR mRNA motifs and interfere with the formation of the cap-dependent initiation complex, resulting in mRNPs with a closed-loop conformation. We demonstrate here that the human DEAD-box protein Rck/p54, which is a component of such complexes and central to P-body assembly, is in considerable molecular excess with respect to cellular mRNAs and enriched to a concentration of 0.5 mM in P-bodies, where it is organized in clusters. Accordingly, multiple binding of p54 proteins along mRNA molecules was detected in vivo. Consistently, the purified protein bound RNA with no sequence specificity and high nanomolar affinity. Moreover, bound RNA molecules had a relaxed conformation. While RNA binding was ATP independent, relaxing of bound RNA was dependent on ATP, though not on its hydrolysis. We propose that Rck/p54 recruitment by sequence-specific translational repressors leads to further binding of Rck/p54 along mRNA molecules, resulting in their masking, unwinding, and ultimately recruitment to P-bodies. Rck/p54 proteins located at the 5' extremity of mRNA can then recruit the decapping complex, thus coupling translational repression and mRNA degradation.
Figures
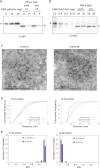

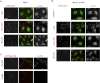
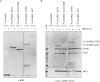
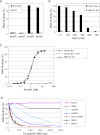

References
-
- Andrey P, Kiêu K, Kress C, Lehmann G, Tirichine L, Liu Z, Biot E, Adenot PG, Hue-Beauvais C, Houba-Herin N, et al. 2010. Statistical analysis of 3D images detects regular spatial distributions of centromeres and chromocenters in animal and plant nuclei. PLoS Comput Biol 6: e1000853 doi: 10.1371/journal.pcbi.1000853 - PMC - PubMed
-
- Ballut L, Marchadier B, Baguet A, Tomasetto C, Seraphin B, Le Hir H 2005. The exon junction core complex is locked onto RNA by inhibition of eIF4AIII ATPase activity. Nat Struct Mol Biol 12: 861–869 - PubMed
-
- Boulton AA, Baker GB, Campagnoni AT, ed. 1990. Molecular neurobiological techniques. In Neuromethods, Vol 16. Humana Press, Clifton, NJ
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
