Tick-borne Nyamanini virus replicates in the nucleus and exhibits unusual genome and matrix protein properties
- PMID: 22837209
- PMCID: PMC3457285
- DOI: 10.1128/JVI.00571-12
Tick-borne Nyamanini virus replicates in the nucleus and exhibits unusual genome and matrix protein properties
Abstract
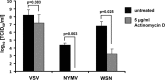
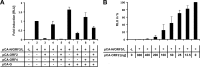
Tick-borne Nyamanini virus (NYMV) is the prototypic member of a recently discovered genus in the order Mononegavirales, designated Nyavirus. The NYMV genome codes for six distinct genes. Sequence similarity and structural properties suggest that genes 1, 5, and 6 encode the nucleoprotein (N), the glycoprotein (G), and the viral polymerase (L), respectively. The function of the other viral genes has been unknown to date. We found that the third NYMV gene codes for a protein which, when coexpressed with N and L, can reconstitute viral polymerase activity, suggesting that it represents a polymerase cofactor. The second viral gene codes for a small protein that inhibits viral polymerase activity and further strongly enhances the formation of virus-like particles when coexpressed with gene 4 and the viral glycoprotein G. This suggests that two distinct proteins serve a matrix protein function in NYMV as previously described for members of the family Filoviridae. We further found that NYMV replicates in the nucleus of infected cells like members of the family Bornaviridae. NYMV is a poor inducer of beta interferon, presumably because the viral genome is 5' monophosphorylated and has a protruding 3' terminus as observed for bornaviruses. Taken together, our results demonstrate that NYMV possesses biological properties previously regarded as typical for filoviruses and bornaviruses, respectively.
Figures



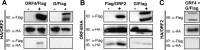


References
-
- Ackermann A, Kugel D, Schneider U, Staeheli P. 2007. Enhanced polymerase activity confers replication competence of Borna disease virus in mice. J. Gen. Virol. 88:3130–3132 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

