Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir
- PMID: 22837215
- PMCID: PMC3457319
- DOI: 10.1128/JVI.01498-12
Eastern chimpanzees, but not bonobos, represent a simian immunodeficiency virus reservoir
Abstract
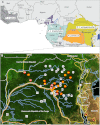
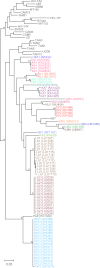
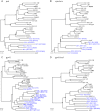
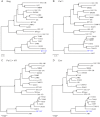
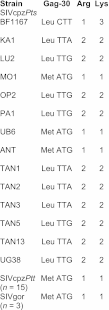
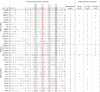
Chimpanzees in west central Africa (Pan troglodytes troglodytes) are endemically infected with simian immunodeficiency viruses (SIVcpzPtt) that have crossed the species barrier to humans and gorillas on at least five occasions, generating pandemic and nonpandemic forms of human immunodeficiency virus type 1 (HIV-1) as well as gorilla SIV (SIVgor). Chimpanzees in east Africa (Pan troglodytes schweinfurthii) are also infected with SIVcpz; however, their viruses (SIVcpzPts) have never been found in humans. To examine whether this is due to a paucity of natural infections, we used noninvasive methods to screen wild-living eastern chimpanzees in the Democratic Republic of the Congo (DRC), Uganda, and Rwanda. We also screened bonobos (Pan paniscus) in the DRC, a species not previously tested for SIV in the wild. Fecal samples (n = 3,108) were collected at 50 field sites, tested for species and subspecies origin, and screened for SIVcpz antibodies and nucleic acids. Of 2,565 samples from eastern chimpanzees, 323 were antibody positive and 92 contained viral RNA. The antibody-positive samples represented 76 individuals from 19 field sites, all sampled north of the Congo River in an area spanning 250,000 km(2). In this region, SIVcpzPts was common and widespread, with seven field sites exhibiting infection rates of 30% or greater. The overall prevalence of SIVcpzPts infection was 13.4% (95% confidence interval, 10.7% to 16.5%). In contrast, none of the 543 bonobo samples from six sites was antibody positive. All newly identified SIVcpzPts strains clustered in strict accordance to their subspecies origin; however, they exhibited considerable genetic diversity, especially in protein domains known to be under strong host selection pressure. Thus, the absence of SIVcpzPts zoonoses cannot be explained by an insufficient primate reservoir. Instead, greater adaptive hurdles may have prevented the successful colonization of humans by P. t. schweinfurthii viruses.
Figures

References
-
- Abascal F, Zardoya R, Posada D. 2005. ProtTest: selection of best-fit models of protein evolution. Bioinformatics 21:2104–2105 - PubMed
-
- Bailes E, et al. 2003. Hybrid origin of SIV in chimpanzees. Science 300:1713. - PubMed
-
- Bennett EL, et al. 2007. Hunting for consensus: reconciling bushmeat harvest, conservation, and development policy in West and Central Africa. Conserv. Biol. 21:884–887 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

