Toward the prediction of FBPase inhibitory activity using chemoinformatic methods
- PMID: 22837677
- PMCID: PMC3397509
- DOI: 10.3390/ijms13067015
Toward the prediction of FBPase inhibitory activity using chemoinformatic methods
Abstract
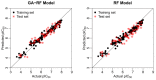
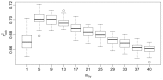
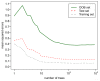
Currently, Chemoinformatic methods are used to perform the prediction for FBPase inhibitory activity. A genetic algorithm-random forest coupled method (GA-RF) was proposed to predict fructose 1,6-bisphosphatase (FBPase) inhibitors to treat type 2 diabetes mellitus using the Mold(2) molecular descriptors. A data set of 126 oxazole and thiazole analogs was used to derive the GA-RF model, yielding the significant non-cross-validated correlation coefficient r(2) (ncv) and cross-validated r(2) (cv) values of 0.96 and 0.67 for the training set, respectively. The statistically significant model was validated by a test set of 64 compounds, producing the prediction correlation coefficient r(2) (pred) of 0.90. More importantly, the building GA-RF model also passed through various criteria suggested by Tropsha and Roy with r(2) (o) and r(2) (m) values of 0.90 and 0.83, respectively. In order to compare with the GA-RF model, a pure RF model developed based on the full descriptors was performed as well for the same data set. The resulting GA-RF model with significantly internal and external prediction capacities is beneficial to the prediction of potential oxazole and thiazole series of FBPase inhibitors prior to chemical synthesis in drug discovery programs.
Keywords: FBPase inhibitor; chemoinformatics methods; genetic algorithm; random forest.
Figures


References
-
- King H., Aubert R.E., Herman W.H. Global burden of diabetes, 1995–2025: Prevalence, numerical estimates, and projections. Diabetes Care. 1998;21:1414–1431. - PubMed
-
- Zhang B.B., Moller D.E. New approaches in the treatment of type 2 diabetes. Curr. Opin. Chem. Biol. 2000;4:4614–4667. - PubMed
-
- Reaven G.M. Pathophysiology of insulin resistance in human disease. Physiol. Rev. 1995;75:4734–4786. - PubMed
-
- Haffner S.M., Lehto S., Rönnemaa T., Pyörälä K., Laakso M. Mortality from coronary heart disease in subjects with type 2 diabetes and in nondiabetic subjects with and without prior myocardial infarction. N. Engl. J. Med. 1998;339:2292–2234. - PubMed
-
- Moller D.E. New drug targets for type 2 diabetes and the metabolic syndrome. Nature. 2001;414:8218–8227. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

